
Maribacter sp. (strain HTCC2170 / KCCM 42371)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Maribacter; unclassified Maribacter
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
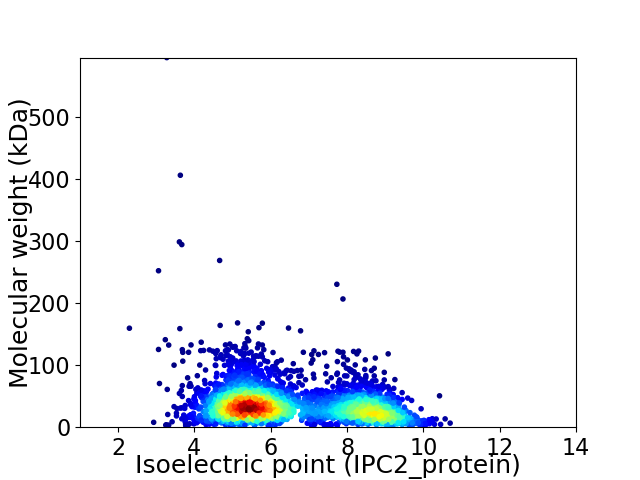
Virtual 2D-PAGE plot for 3411 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
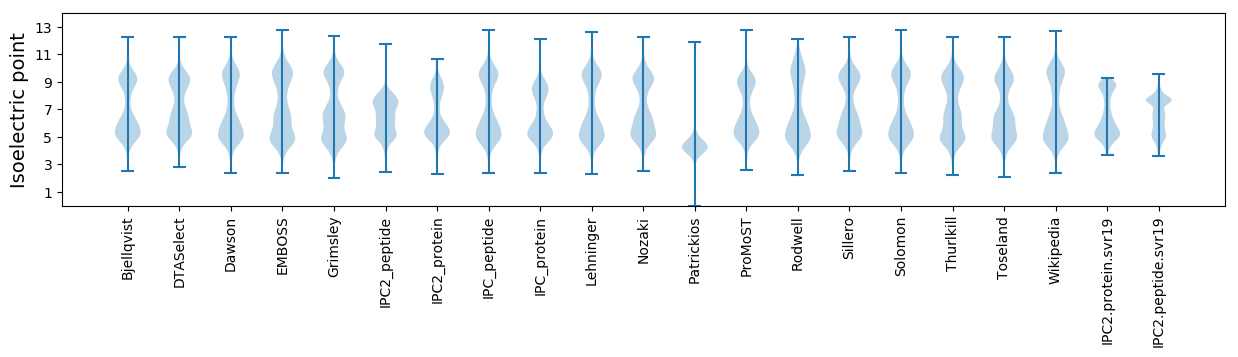
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4AT64|A4AT64_MARSH Uncharacterized protein OS=Maribacter sp. (strain HTCC2170 / KCCM 42371) OX=313603 GN=FB2170_16156 PE=4 SV=1
MM1 pKa = 7.65RR2 pKa = 11.84RR3 pKa = 11.84NSQSFLSKK11 pKa = 9.81LTFGIILSLLIFYY24 pKa = 10.45HH25 pKa = 7.01GEE27 pKa = 4.13TFSQTTFTEE36 pKa = 4.1SAAAYY41 pKa = 9.39GLNLGGNKK49 pKa = 10.19DD50 pKa = 4.28GGHH53 pKa = 6.48AWADD57 pKa = 3.38YY58 pKa = 11.38DD59 pKa = 5.18LDD61 pKa = 4.87GDD63 pKa = 3.98FDD65 pKa = 5.18LVVNTQGNGKK75 pKa = 10.14LYY77 pKa = 10.68RR78 pKa = 11.84NDD80 pKa = 3.48GGSFTDD86 pKa = 3.38VTGTLAPDD94 pKa = 4.04FNGGSLEE101 pKa = 4.09RR102 pKa = 11.84TVLFVDD108 pKa = 4.95FNNDD112 pKa = 2.65GYY114 pKa = 10.47PDD116 pKa = 3.59VFRR119 pKa = 11.84NKK121 pKa = 10.39HH122 pKa = 5.12NDD124 pKa = 2.95LRR126 pKa = 11.84IYY128 pKa = 10.64LQDD131 pKa = 3.61PATNRR136 pKa = 11.84FGNGTGGTVPNQRR149 pKa = 11.84FTSMTDD155 pKa = 3.01GMNTEE160 pKa = 4.4GAGALDD166 pKa = 3.82YY167 pKa = 11.58DD168 pKa = 4.48GDD170 pKa = 4.06GDD172 pKa = 5.35LDD174 pKa = 4.52LFVDD178 pKa = 3.6NHH180 pKa = 6.08NFGIDD185 pKa = 2.86ILQNDD190 pKa = 3.94GTGNFTHH197 pKa = 6.0VTRR200 pKa = 11.84KK201 pKa = 9.87ADD203 pKa = 3.86SPNPPYY209 pKa = 10.76DD210 pKa = 3.69VLNSTTWPLGLVQDD224 pKa = 4.57ATDD227 pKa = 3.75GDD229 pKa = 4.43YY230 pKa = 11.73GSATDD235 pKa = 4.66FNDD238 pKa = 4.31DD239 pKa = 3.06GWVDD243 pKa = 2.99IVVRR247 pKa = 11.84KK248 pKa = 9.7RR249 pKa = 11.84DD250 pKa = 3.67QVDD253 pKa = 3.68LFTNVGGTFQNGVNIDD269 pKa = 3.74DD270 pKa = 4.7ANNNNKK276 pKa = 9.7GAVAFYY282 pKa = 11.03DD283 pKa = 3.87FDD285 pKa = 4.46NDD287 pKa = 4.64GDD289 pKa = 4.1FDD291 pKa = 4.28MYY293 pKa = 8.96WTEE296 pKa = 3.79NGNNQIHH303 pKa = 6.81RR304 pKa = 11.84NNGDD308 pKa = 3.63GTWTGLGAATGIPISFSGQIEE329 pKa = 4.09GLACGDD335 pKa = 3.56VDD337 pKa = 4.26NDD339 pKa = 3.4GDD341 pKa = 4.12IDD343 pKa = 3.65IFLAGGANKK352 pKa = 10.33LFLNQINNGGGAMSFVDD369 pKa = 4.36SGLTFSNSGGEE380 pKa = 4.09GCTFIDD386 pKa = 3.97IDD388 pKa = 4.23QDD390 pKa = 3.5GDD392 pKa = 3.58LDD394 pKa = 4.95LYY396 pKa = 9.76TNRR399 pKa = 11.84SGNNRR404 pKa = 11.84LYY406 pKa = 10.64INNLMANQANHH417 pKa = 6.79LYY419 pKa = 10.16IDD421 pKa = 3.48IVEE424 pKa = 4.59DD425 pKa = 3.59RR426 pKa = 11.84DD427 pKa = 3.81AFGLINTEE435 pKa = 3.63EE436 pKa = 4.38RR437 pKa = 11.84FGVGATAKK445 pKa = 10.58ILDD448 pKa = 4.02CDD450 pKa = 4.14GNVISGTRR458 pKa = 11.84EE459 pKa = 3.64VNGGFGHH466 pKa = 6.6GTQGPGRR473 pKa = 11.84IHH475 pKa = 6.97FGLPSGPLTPIVVEE489 pKa = 4.01VSFPRR494 pKa = 11.84TASGRR499 pKa = 11.84VVVRR503 pKa = 11.84QQLIPNDD510 pKa = 3.84YY511 pKa = 11.13NNGSINLLDD520 pKa = 4.11VFPDD524 pKa = 3.77SANQPPTATDD534 pKa = 3.35EE535 pKa = 4.33YY536 pKa = 8.47TTVVEE541 pKa = 4.25NSFVSLDD548 pKa = 3.69PLIDD552 pKa = 3.8NGNGADD558 pKa = 4.14SDD560 pKa = 4.48PEE562 pKa = 4.34GEE564 pKa = 4.08PLEE567 pKa = 4.74VISVTQPSNGVSVLNGDD584 pKa = 3.57GTVTYY589 pKa = 9.85TPTTGFFGNDD599 pKa = 2.6SFTYY603 pKa = 9.73TMRR606 pKa = 11.84DD607 pKa = 3.17NANCTFTSEE616 pKa = 4.11EE617 pKa = 4.2ATGSIYY623 pKa = 9.65ITVFPDD629 pKa = 3.14SDD631 pKa = 3.84NDD633 pKa = 3.91TVADD637 pKa = 4.9RR638 pKa = 11.84VDD640 pKa = 4.93LDD642 pKa = 4.5DD643 pKa = 6.95DD644 pKa = 4.5NDD646 pKa = 4.03GLLDD650 pKa = 4.24ADD652 pKa = 4.5EE653 pKa = 5.37LNTIISNSQQDD664 pKa = 4.01CTGEE668 pKa = 4.13TTLDD672 pKa = 3.6FSSAATLVSGTALQLGAVYY691 pKa = 10.25RR692 pKa = 11.84ISNITTGTDD701 pKa = 2.97ALVTIDD707 pKa = 3.3QTFNATVANIDD718 pKa = 3.74NNGSEE723 pKa = 4.27PASFRR728 pKa = 11.84PQTAFNLTNIGDD740 pKa = 3.55QGYY743 pKa = 9.61IEE745 pKa = 4.29YY746 pKa = 10.35RR747 pKa = 11.84IQFVNSGGSAPVVISKK763 pKa = 9.35FFTNFNDD770 pKa = 3.18IDD772 pKa = 3.99GNSNYY777 pKa = 10.27GEE779 pKa = 4.55QIWSDD784 pKa = 3.43NPTSYY789 pKa = 10.47IISNPTEE796 pKa = 3.96LTMSTDD802 pKa = 3.7GSWVLGTAGINEE814 pKa = 4.17FPGAGNTFPEE824 pKa = 4.2VNYY827 pKa = 10.13GVNYY831 pKa = 10.17NSQSEE836 pKa = 3.82ISIRR840 pKa = 11.84VGAVARR846 pKa = 11.84VAGASASGRR855 pKa = 11.84QHH857 pKa = 6.64NIEE860 pKa = 4.51FSCLTNYY867 pKa = 9.38VNPEE871 pKa = 3.95TYY873 pKa = 10.77GIDD876 pKa = 3.55SDD878 pKa = 4.15SDD880 pKa = 3.95GIANHH885 pKa = 7.19LDD887 pKa = 3.6LDD889 pKa = 4.02SDD891 pKa = 3.75NDD893 pKa = 4.55GIYY896 pKa = 10.79DD897 pKa = 3.6AVEE900 pKa = 4.29AGHH903 pKa = 5.97SQSHH907 pKa = 6.05SNGVVNGPYY916 pKa = 9.94GANGLANVVEE926 pKa = 4.67TAAEE930 pKa = 4.07SGAINYY936 pKa = 7.6TILDD940 pKa = 3.63TDD942 pKa = 3.76GTDD945 pKa = 3.41PADD948 pKa = 4.76FLDD951 pKa = 4.35TDD953 pKa = 4.45SDD955 pKa = 4.97DD956 pKa = 5.67DD957 pKa = 4.24GCSDD961 pKa = 5.06ANEE964 pKa = 4.77AYY966 pKa = 10.92NNANADD972 pKa = 3.64GGDD975 pKa = 3.49NEE977 pKa = 5.0YY978 pKa = 10.86YY979 pKa = 10.05ATGNPPATDD988 pKa = 3.11GFGRR992 pKa = 11.84VTGATYY998 pKa = 9.05PVPADD1003 pKa = 3.21IDD1005 pKa = 3.63TDD1007 pKa = 3.61STYY1010 pKa = 11.92DD1011 pKa = 3.38HH1012 pKa = 7.18LEE1014 pKa = 3.93VGLAPSISTQPPNTNVCPGCDD1035 pKa = 2.81ITIEE1039 pKa = 4.36VIASNSDD1046 pKa = 3.31TYY1048 pKa = 11.04QWQLDD1053 pKa = 4.14DD1054 pKa = 3.92GSGWVDD1060 pKa = 3.52LTDD1063 pKa = 3.3SGIYY1067 pKa = 9.95SDD1069 pKa = 4.14TSTSILTITNATAPDD1084 pKa = 3.57NGNEE1088 pKa = 3.72YY1089 pKa = 10.52RR1090 pKa = 11.84VIVSNTMFICSSEE1103 pKa = 4.19TSNSAVLTLRR1113 pKa = 11.84VNTIITNRR1121 pKa = 11.84RR1122 pKa = 11.84ITHH1125 pKa = 6.35RR1126 pKa = 11.84VNKK1129 pKa = 9.94NN1130 pKa = 2.67
MM1 pKa = 7.65RR2 pKa = 11.84RR3 pKa = 11.84NSQSFLSKK11 pKa = 9.81LTFGIILSLLIFYY24 pKa = 10.45HH25 pKa = 7.01GEE27 pKa = 4.13TFSQTTFTEE36 pKa = 4.1SAAAYY41 pKa = 9.39GLNLGGNKK49 pKa = 10.19DD50 pKa = 4.28GGHH53 pKa = 6.48AWADD57 pKa = 3.38YY58 pKa = 11.38DD59 pKa = 5.18LDD61 pKa = 4.87GDD63 pKa = 3.98FDD65 pKa = 5.18LVVNTQGNGKK75 pKa = 10.14LYY77 pKa = 10.68RR78 pKa = 11.84NDD80 pKa = 3.48GGSFTDD86 pKa = 3.38VTGTLAPDD94 pKa = 4.04FNGGSLEE101 pKa = 4.09RR102 pKa = 11.84TVLFVDD108 pKa = 4.95FNNDD112 pKa = 2.65GYY114 pKa = 10.47PDD116 pKa = 3.59VFRR119 pKa = 11.84NKK121 pKa = 10.39HH122 pKa = 5.12NDD124 pKa = 2.95LRR126 pKa = 11.84IYY128 pKa = 10.64LQDD131 pKa = 3.61PATNRR136 pKa = 11.84FGNGTGGTVPNQRR149 pKa = 11.84FTSMTDD155 pKa = 3.01GMNTEE160 pKa = 4.4GAGALDD166 pKa = 3.82YY167 pKa = 11.58DD168 pKa = 4.48GDD170 pKa = 4.06GDD172 pKa = 5.35LDD174 pKa = 4.52LFVDD178 pKa = 3.6NHH180 pKa = 6.08NFGIDD185 pKa = 2.86ILQNDD190 pKa = 3.94GTGNFTHH197 pKa = 6.0VTRR200 pKa = 11.84KK201 pKa = 9.87ADD203 pKa = 3.86SPNPPYY209 pKa = 10.76DD210 pKa = 3.69VLNSTTWPLGLVQDD224 pKa = 4.57ATDD227 pKa = 3.75GDD229 pKa = 4.43YY230 pKa = 11.73GSATDD235 pKa = 4.66FNDD238 pKa = 4.31DD239 pKa = 3.06GWVDD243 pKa = 2.99IVVRR247 pKa = 11.84KK248 pKa = 9.7RR249 pKa = 11.84DD250 pKa = 3.67QVDD253 pKa = 3.68LFTNVGGTFQNGVNIDD269 pKa = 3.74DD270 pKa = 4.7ANNNNKK276 pKa = 9.7GAVAFYY282 pKa = 11.03DD283 pKa = 3.87FDD285 pKa = 4.46NDD287 pKa = 4.64GDD289 pKa = 4.1FDD291 pKa = 4.28MYY293 pKa = 8.96WTEE296 pKa = 3.79NGNNQIHH303 pKa = 6.81RR304 pKa = 11.84NNGDD308 pKa = 3.63GTWTGLGAATGIPISFSGQIEE329 pKa = 4.09GLACGDD335 pKa = 3.56VDD337 pKa = 4.26NDD339 pKa = 3.4GDD341 pKa = 4.12IDD343 pKa = 3.65IFLAGGANKK352 pKa = 10.33LFLNQINNGGGAMSFVDD369 pKa = 4.36SGLTFSNSGGEE380 pKa = 4.09GCTFIDD386 pKa = 3.97IDD388 pKa = 4.23QDD390 pKa = 3.5GDD392 pKa = 3.58LDD394 pKa = 4.95LYY396 pKa = 9.76TNRR399 pKa = 11.84SGNNRR404 pKa = 11.84LYY406 pKa = 10.64INNLMANQANHH417 pKa = 6.79LYY419 pKa = 10.16IDD421 pKa = 3.48IVEE424 pKa = 4.59DD425 pKa = 3.59RR426 pKa = 11.84DD427 pKa = 3.81AFGLINTEE435 pKa = 3.63EE436 pKa = 4.38RR437 pKa = 11.84FGVGATAKK445 pKa = 10.58ILDD448 pKa = 4.02CDD450 pKa = 4.14GNVISGTRR458 pKa = 11.84EE459 pKa = 3.64VNGGFGHH466 pKa = 6.6GTQGPGRR473 pKa = 11.84IHH475 pKa = 6.97FGLPSGPLTPIVVEE489 pKa = 4.01VSFPRR494 pKa = 11.84TASGRR499 pKa = 11.84VVVRR503 pKa = 11.84QQLIPNDD510 pKa = 3.84YY511 pKa = 11.13NNGSINLLDD520 pKa = 4.11VFPDD524 pKa = 3.77SANQPPTATDD534 pKa = 3.35EE535 pKa = 4.33YY536 pKa = 8.47TTVVEE541 pKa = 4.25NSFVSLDD548 pKa = 3.69PLIDD552 pKa = 3.8NGNGADD558 pKa = 4.14SDD560 pKa = 4.48PEE562 pKa = 4.34GEE564 pKa = 4.08PLEE567 pKa = 4.74VISVTQPSNGVSVLNGDD584 pKa = 3.57GTVTYY589 pKa = 9.85TPTTGFFGNDD599 pKa = 2.6SFTYY603 pKa = 9.73TMRR606 pKa = 11.84DD607 pKa = 3.17NANCTFTSEE616 pKa = 4.11EE617 pKa = 4.2ATGSIYY623 pKa = 9.65ITVFPDD629 pKa = 3.14SDD631 pKa = 3.84NDD633 pKa = 3.91TVADD637 pKa = 4.9RR638 pKa = 11.84VDD640 pKa = 4.93LDD642 pKa = 4.5DD643 pKa = 6.95DD644 pKa = 4.5NDD646 pKa = 4.03GLLDD650 pKa = 4.24ADD652 pKa = 4.5EE653 pKa = 5.37LNTIISNSQQDD664 pKa = 4.01CTGEE668 pKa = 4.13TTLDD672 pKa = 3.6FSSAATLVSGTALQLGAVYY691 pKa = 10.25RR692 pKa = 11.84ISNITTGTDD701 pKa = 2.97ALVTIDD707 pKa = 3.3QTFNATVANIDD718 pKa = 3.74NNGSEE723 pKa = 4.27PASFRR728 pKa = 11.84PQTAFNLTNIGDD740 pKa = 3.55QGYY743 pKa = 9.61IEE745 pKa = 4.29YY746 pKa = 10.35RR747 pKa = 11.84IQFVNSGGSAPVVISKK763 pKa = 9.35FFTNFNDD770 pKa = 3.18IDD772 pKa = 3.99GNSNYY777 pKa = 10.27GEE779 pKa = 4.55QIWSDD784 pKa = 3.43NPTSYY789 pKa = 10.47IISNPTEE796 pKa = 3.96LTMSTDD802 pKa = 3.7GSWVLGTAGINEE814 pKa = 4.17FPGAGNTFPEE824 pKa = 4.2VNYY827 pKa = 10.13GVNYY831 pKa = 10.17NSQSEE836 pKa = 3.82ISIRR840 pKa = 11.84VGAVARR846 pKa = 11.84VAGASASGRR855 pKa = 11.84QHH857 pKa = 6.64NIEE860 pKa = 4.51FSCLTNYY867 pKa = 9.38VNPEE871 pKa = 3.95TYY873 pKa = 10.77GIDD876 pKa = 3.55SDD878 pKa = 4.15SDD880 pKa = 3.95GIANHH885 pKa = 7.19LDD887 pKa = 3.6LDD889 pKa = 4.02SDD891 pKa = 3.75NDD893 pKa = 4.55GIYY896 pKa = 10.79DD897 pKa = 3.6AVEE900 pKa = 4.29AGHH903 pKa = 5.97SQSHH907 pKa = 6.05SNGVVNGPYY916 pKa = 9.94GANGLANVVEE926 pKa = 4.67TAAEE930 pKa = 4.07SGAINYY936 pKa = 7.6TILDD940 pKa = 3.63TDD942 pKa = 3.76GTDD945 pKa = 3.41PADD948 pKa = 4.76FLDD951 pKa = 4.35TDD953 pKa = 4.45SDD955 pKa = 4.97DD956 pKa = 5.67DD957 pKa = 4.24GCSDD961 pKa = 5.06ANEE964 pKa = 4.77AYY966 pKa = 10.92NNANADD972 pKa = 3.64GGDD975 pKa = 3.49NEE977 pKa = 5.0YY978 pKa = 10.86YY979 pKa = 10.05ATGNPPATDD988 pKa = 3.11GFGRR992 pKa = 11.84VTGATYY998 pKa = 9.05PVPADD1003 pKa = 3.21IDD1005 pKa = 3.63TDD1007 pKa = 3.61STYY1010 pKa = 11.92DD1011 pKa = 3.38HH1012 pKa = 7.18LEE1014 pKa = 3.93VGLAPSISTQPPNTNVCPGCDD1035 pKa = 2.81ITIEE1039 pKa = 4.36VIASNSDD1046 pKa = 3.31TYY1048 pKa = 11.04QWQLDD1053 pKa = 4.14DD1054 pKa = 3.92GSGWVDD1060 pKa = 3.52LTDD1063 pKa = 3.3SGIYY1067 pKa = 9.95SDD1069 pKa = 4.14TSTSILTITNATAPDD1084 pKa = 3.57NGNEE1088 pKa = 3.72YY1089 pKa = 10.52RR1090 pKa = 11.84VIVSNTMFICSSEE1103 pKa = 4.19TSNSAVLTLRR1113 pKa = 11.84VNTIITNRR1121 pKa = 11.84RR1122 pKa = 11.84ITHH1125 pKa = 6.35RR1126 pKa = 11.84VNKK1129 pKa = 9.94NN1130 pKa = 2.67
Molecular weight: 120.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4AUD4|A4AUD4_MARSH Uncharacterized protein OS=Maribacter sp. (strain HTCC2170 / KCCM 42371) OX=313603 GN=FB2170_08769 PE=4 SV=1
MM1 pKa = 7.62PKK3 pKa = 10.27SNRR6 pKa = 11.84SRR8 pKa = 11.84SWIDD12 pKa = 2.94ILSEE16 pKa = 4.26LIEE19 pKa = 4.31VFFKK23 pKa = 10.97SAGRR27 pKa = 11.84SKK29 pKa = 10.72RR30 pKa = 11.84GRR32 pKa = 11.84TWHH35 pKa = 4.78QHH37 pKa = 3.44VVPYY41 pKa = 10.44EE42 pKa = 4.27DD43 pKa = 3.53DD44 pKa = 3.27WAVRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 4.02GNKK53 pKa = 10.25RR54 pKa = 11.84ITSKK58 pKa = 10.32HH59 pKa = 5.07RR60 pKa = 11.84RR61 pKa = 11.84QDD63 pKa = 3.24TAIRR67 pKa = 11.84KK68 pKa = 9.06AKK70 pKa = 9.3TLARR74 pKa = 11.84KK75 pKa = 9.3HH76 pKa = 5.81KK77 pKa = 10.31ADD79 pKa = 4.04VIIHH83 pKa = 6.58RR84 pKa = 11.84SDD86 pKa = 2.73GTIRR90 pKa = 11.84DD91 pKa = 4.15RR92 pKa = 11.84INYY95 pKa = 8.01DD96 pKa = 2.82
MM1 pKa = 7.62PKK3 pKa = 10.27SNRR6 pKa = 11.84SRR8 pKa = 11.84SWIDD12 pKa = 2.94ILSEE16 pKa = 4.26LIEE19 pKa = 4.31VFFKK23 pKa = 10.97SAGRR27 pKa = 11.84SKK29 pKa = 10.72RR30 pKa = 11.84GRR32 pKa = 11.84TWHH35 pKa = 4.78QHH37 pKa = 3.44VVPYY41 pKa = 10.44EE42 pKa = 4.27DD43 pKa = 3.53DD44 pKa = 3.27WAVRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 4.02GNKK53 pKa = 10.25RR54 pKa = 11.84ITSKK58 pKa = 10.32HH59 pKa = 5.07RR60 pKa = 11.84RR61 pKa = 11.84QDD63 pKa = 3.24TAIRR67 pKa = 11.84KK68 pKa = 9.06AKK70 pKa = 9.3TLARR74 pKa = 11.84KK75 pKa = 9.3HH76 pKa = 5.81KK77 pKa = 10.31ADD79 pKa = 4.04VIIHH83 pKa = 6.58RR84 pKa = 11.84SDD86 pKa = 2.73GTIRR90 pKa = 11.84DD91 pKa = 4.15RR92 pKa = 11.84INYY95 pKa = 8.01DD96 pKa = 2.82
Molecular weight: 11.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1168930 |
22 |
5669 |
342.7 |
38.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.265 ± 0.041 | 0.75 ± 0.017 |
5.794 ± 0.048 | 6.67 ± 0.046 |
5.117 ± 0.037 | 6.937 ± 0.066 |
1.874 ± 0.024 | 7.727 ± 0.041 |
7.626 ± 0.068 | 9.23 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.401 ± 0.025 | 5.946 ± 0.038 |
3.555 ± 0.029 | 3.18 ± 0.021 |
3.508 ± 0.031 | 6.484 ± 0.031 |
5.552 ± 0.063 | 6.286 ± 0.033 |
1.177 ± 0.018 | 3.92 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
