
Roseburia sp. 499
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Roseburia; unclassified Roseburia
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
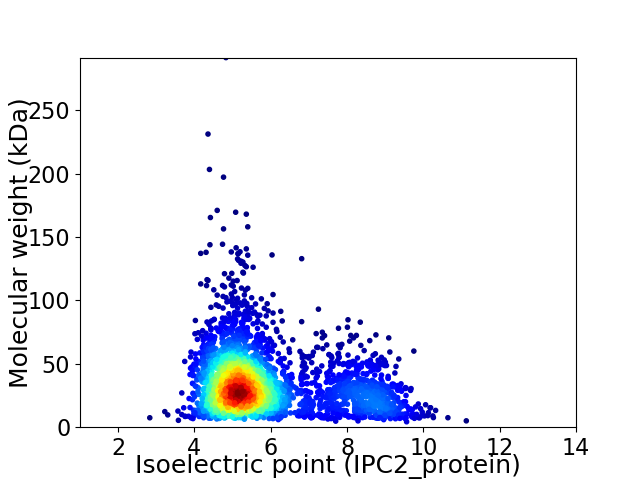
Virtual 2D-PAGE plot for 3105 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
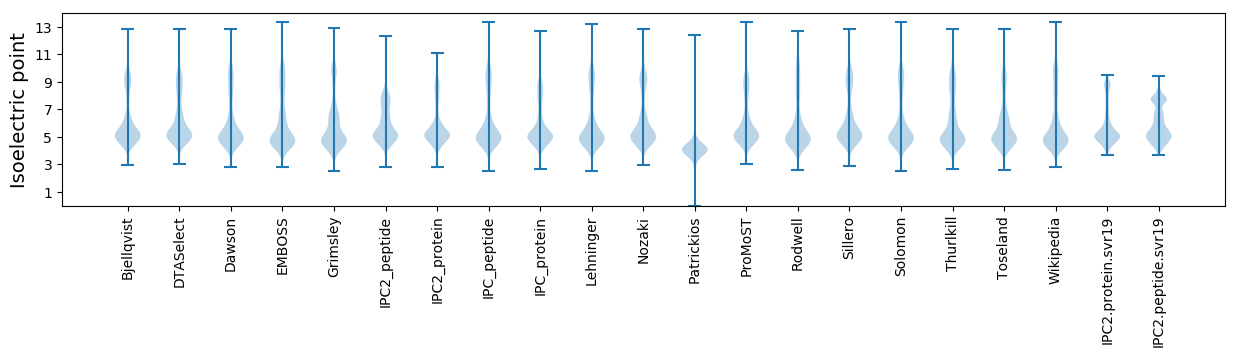
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q9JZD8|A0A1Q9JZD8_9FIRM GntR family transcriptional regulator OS=Roseburia sp. 499 OX=1261634 GN=BIV20_04880 PE=4 SV=1
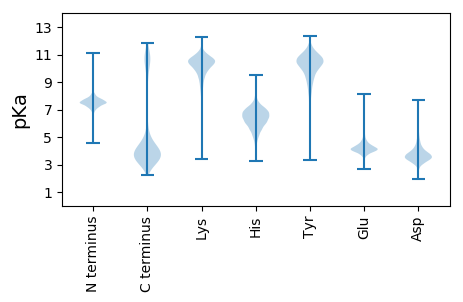
MM1 pKa = 7.22KK2 pKa = 10.23RR3 pKa = 11.84KK4 pKa = 9.3LVSLMLVAAMAASMVVGCGNNSNDD28 pKa = 4.42KK29 pKa = 10.98GSNDD33 pKa = 3.21KK34 pKa = 11.34GKK36 pKa = 8.37TEE38 pKa = 3.89EE39 pKa = 4.29AGGEE43 pKa = 4.03KK44 pKa = 10.48DD45 pKa = 3.6EE46 pKa = 5.0SGSTASGDD54 pKa = 3.24GRR56 pKa = 11.84VYY58 pKa = 11.19LLNFKK63 pKa = 10.65PEE65 pKa = 4.02TDD67 pKa = 4.09QAWQDD72 pKa = 3.57LADD75 pKa = 3.84VYY77 pKa = 11.05TDD79 pKa = 3.39EE80 pKa = 4.89TGVEE84 pKa = 4.43VNVLTAADD92 pKa = 4.09GQYY95 pKa = 9.12STTMQSEE102 pKa = 4.35MAKK105 pKa = 10.63DD106 pKa = 3.41EE107 pKa = 4.5APTIFNIGNTTAAQTWNDD125 pKa = 3.39YY126 pKa = 9.18TLDD129 pKa = 3.91LKK131 pKa = 11.18DD132 pKa = 3.62SEE134 pKa = 4.94LYY136 pKa = 10.46KK137 pKa = 10.9HH138 pKa = 6.18LTDD141 pKa = 3.83KK142 pKa = 11.03SLSITYY148 pKa = 9.37DD149 pKa = 3.23GKK151 pKa = 10.26IAAVANCYY159 pKa = 8.88EE160 pKa = 4.63CYY162 pKa = 10.64GIIYY166 pKa = 9.51NKK168 pKa = 10.3KK169 pKa = 9.51ILNDD173 pKa = 3.36YY174 pKa = 7.56CTLDD178 pKa = 3.54GAVIASPDD186 pKa = 3.94EE187 pKa = 3.92ITSFDD192 pKa = 3.54TLKK195 pKa = 11.08AVAEE199 pKa = 4.77DD200 pKa = 3.05INARR204 pKa = 11.84VDD206 pKa = 3.91EE207 pKa = 4.69INDD210 pKa = 3.27EE211 pKa = 4.38FGYY214 pKa = 10.42EE215 pKa = 3.89LQGAFASAGLDD226 pKa = 3.32SGSSWRR232 pKa = 11.84FSGHH236 pKa = 6.45LANMPLYY243 pKa = 11.0YY244 pKa = 10.18EE245 pKa = 4.8FKK247 pKa = 10.8DD248 pKa = 4.85DD249 pKa = 5.15GCDD252 pKa = 3.98LINGEE257 pKa = 4.14ATIDD261 pKa = 3.57GTYY264 pKa = 10.34LDD266 pKa = 3.82QYY268 pKa = 9.78KK269 pKa = 10.42AVWDD273 pKa = 4.42LYY275 pKa = 10.95VNTSGADD282 pKa = 3.45PKK284 pKa = 10.21TLNSGALNAEE294 pKa = 4.44SEE296 pKa = 4.48FGMEE300 pKa = 4.02EE301 pKa = 3.67AVFYY305 pKa = 11.31QNGDD309 pKa = 3.31WEE311 pKa = 4.58FSPLTSDD318 pKa = 3.23EE319 pKa = 3.96NGYY322 pKa = 10.35LVTADD327 pKa = 5.35DD328 pKa = 4.01IGMMPIYY335 pKa = 10.46FGVDD339 pKa = 3.35DD340 pKa = 4.94EE341 pKa = 4.57NQGLCVGTEE350 pKa = 4.14NYY352 pKa = 8.28WAVNSQASQEE362 pKa = 4.92DD363 pKa = 3.4IDD365 pKa = 3.84ATLAFLEE372 pKa = 4.41WVITSDD378 pKa = 3.41EE379 pKa = 4.07GRR381 pKa = 11.84DD382 pKa = 3.63AITNTMGLSAPYY394 pKa = 8.83DD395 pKa = 3.92TFTGDD400 pKa = 3.52YY401 pKa = 9.52EE402 pKa = 4.37SANAFVQDD410 pKa = 3.87SNALMSAGKK419 pKa = 8.67TSVAWSFNATPNVDD433 pKa = 3.0DD434 pKa = 3.57WRR436 pKa = 11.84ADD438 pKa = 3.45VVSALTAYY446 pKa = 9.73TDD448 pKa = 4.83GSGDD452 pKa = 3.09WDD454 pKa = 3.61AVKK457 pKa = 9.09TAFVDD462 pKa = 4.35GWANQWTLAHH472 pKa = 7.02EE473 pKa = 5.36DD474 pKa = 3.71DD475 pKa = 4.31AQQ477 pKa = 4.77
MM1 pKa = 7.22KK2 pKa = 10.23RR3 pKa = 11.84KK4 pKa = 9.3LVSLMLVAAMAASMVVGCGNNSNDD28 pKa = 4.42KK29 pKa = 10.98GSNDD33 pKa = 3.21KK34 pKa = 11.34GKK36 pKa = 8.37TEE38 pKa = 3.89EE39 pKa = 4.29AGGEE43 pKa = 4.03KK44 pKa = 10.48DD45 pKa = 3.6EE46 pKa = 5.0SGSTASGDD54 pKa = 3.24GRR56 pKa = 11.84VYY58 pKa = 11.19LLNFKK63 pKa = 10.65PEE65 pKa = 4.02TDD67 pKa = 4.09QAWQDD72 pKa = 3.57LADD75 pKa = 3.84VYY77 pKa = 11.05TDD79 pKa = 3.39EE80 pKa = 4.89TGVEE84 pKa = 4.43VNVLTAADD92 pKa = 4.09GQYY95 pKa = 9.12STTMQSEE102 pKa = 4.35MAKK105 pKa = 10.63DD106 pKa = 3.41EE107 pKa = 4.5APTIFNIGNTTAAQTWNDD125 pKa = 3.39YY126 pKa = 9.18TLDD129 pKa = 3.91LKK131 pKa = 11.18DD132 pKa = 3.62SEE134 pKa = 4.94LYY136 pKa = 10.46KK137 pKa = 10.9HH138 pKa = 6.18LTDD141 pKa = 3.83KK142 pKa = 11.03SLSITYY148 pKa = 9.37DD149 pKa = 3.23GKK151 pKa = 10.26IAAVANCYY159 pKa = 8.88EE160 pKa = 4.63CYY162 pKa = 10.64GIIYY166 pKa = 9.51NKK168 pKa = 10.3KK169 pKa = 9.51ILNDD173 pKa = 3.36YY174 pKa = 7.56CTLDD178 pKa = 3.54GAVIASPDD186 pKa = 3.94EE187 pKa = 3.92ITSFDD192 pKa = 3.54TLKK195 pKa = 11.08AVAEE199 pKa = 4.77DD200 pKa = 3.05INARR204 pKa = 11.84VDD206 pKa = 3.91EE207 pKa = 4.69INDD210 pKa = 3.27EE211 pKa = 4.38FGYY214 pKa = 10.42EE215 pKa = 3.89LQGAFASAGLDD226 pKa = 3.32SGSSWRR232 pKa = 11.84FSGHH236 pKa = 6.45LANMPLYY243 pKa = 11.0YY244 pKa = 10.18EE245 pKa = 4.8FKK247 pKa = 10.8DD248 pKa = 4.85DD249 pKa = 5.15GCDD252 pKa = 3.98LINGEE257 pKa = 4.14ATIDD261 pKa = 3.57GTYY264 pKa = 10.34LDD266 pKa = 3.82QYY268 pKa = 9.78KK269 pKa = 10.42AVWDD273 pKa = 4.42LYY275 pKa = 10.95VNTSGADD282 pKa = 3.45PKK284 pKa = 10.21TLNSGALNAEE294 pKa = 4.44SEE296 pKa = 4.48FGMEE300 pKa = 4.02EE301 pKa = 3.67AVFYY305 pKa = 11.31QNGDD309 pKa = 3.31WEE311 pKa = 4.58FSPLTSDD318 pKa = 3.23EE319 pKa = 3.96NGYY322 pKa = 10.35LVTADD327 pKa = 5.35DD328 pKa = 4.01IGMMPIYY335 pKa = 10.46FGVDD339 pKa = 3.35DD340 pKa = 4.94EE341 pKa = 4.57NQGLCVGTEE350 pKa = 4.14NYY352 pKa = 8.28WAVNSQASQEE362 pKa = 4.92DD363 pKa = 3.4IDD365 pKa = 3.84ATLAFLEE372 pKa = 4.41WVITSDD378 pKa = 3.41EE379 pKa = 4.07GRR381 pKa = 11.84DD382 pKa = 3.63AITNTMGLSAPYY394 pKa = 8.83DD395 pKa = 3.92TFTGDD400 pKa = 3.52YY401 pKa = 9.52EE402 pKa = 4.37SANAFVQDD410 pKa = 3.87SNALMSAGKK419 pKa = 8.67TSVAWSFNATPNVDD433 pKa = 3.0DD434 pKa = 3.57WRR436 pKa = 11.84ADD438 pKa = 3.45VVSALTAYY446 pKa = 9.73TDD448 pKa = 4.83GSGDD452 pKa = 3.09WDD454 pKa = 3.61AVKK457 pKa = 9.09TAFVDD462 pKa = 4.35GWANQWTLAHH472 pKa = 7.02EE473 pKa = 5.36DD474 pKa = 3.71DD475 pKa = 4.31AQQ477 pKa = 4.77
Molecular weight: 51.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q9K4F6|A0A1Q9K4F6_9FIRM Tyrosine--tRNA ligase OS=Roseburia sp. 499 OX=1261634 GN=tyrS PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
974933 |
37 |
2567 |
314.0 |
35.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.795 ± 0.04 | 1.435 ± 0.018 |
5.371 ± 0.035 | 8.715 ± 0.06 |
4.05 ± 0.028 | 6.692 ± 0.037 |
1.617 ± 0.018 | 7.745 ± 0.045 |
7.381 ± 0.043 | 8.647 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.265 ± 0.022 | 4.725 ± 0.034 |
2.921 ± 0.021 | 3.541 ± 0.031 |
3.969 ± 0.028 | 5.674 ± 0.035 |
5.275 ± 0.041 | 6.967 ± 0.038 |
0.871 ± 0.015 | 4.343 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
