
Ferrimonas lipolytica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Ferrimonadaceae; Ferrimonas
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
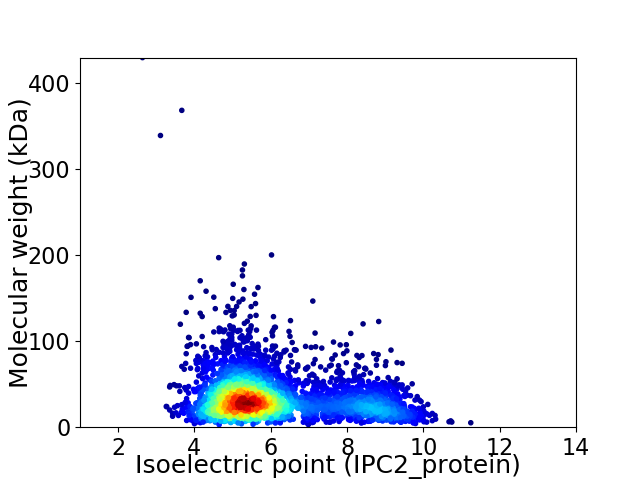
Virtual 2D-PAGE plot for 3539 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
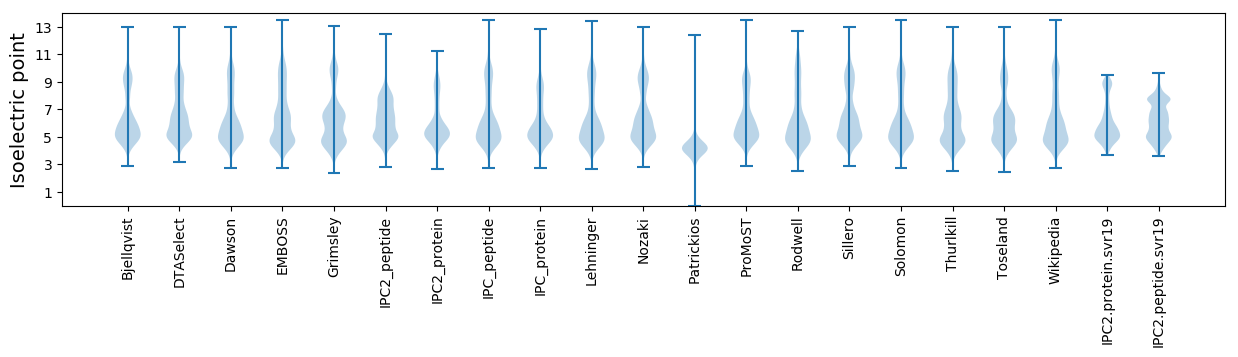
* You can choose from 21 different methods for calculating isoelectric point
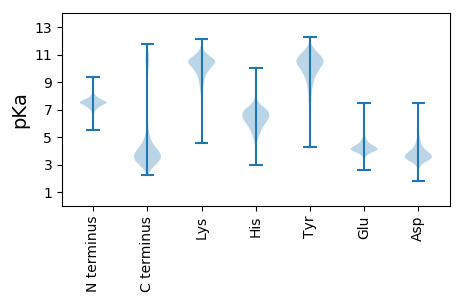
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6H1UBZ8|A0A6H1UBZ8_9GAMM Adenine phosphoribosyltransferase OS=Ferrimonas lipolytica OX=2724191 GN=apt PE=3 SV=1
MM1 pKa = 7.19KK2 pKa = 10.31RR3 pKa = 11.84SLVTLGVIVALAGCDD18 pKa = 3.48SDD20 pKa = 3.54NHH22 pKa = 5.83NVVTSDD28 pKa = 3.34FTLQLLHH35 pKa = 6.33VADD38 pKa = 4.35MDD40 pKa = 4.1GSNATVLEE48 pKa = 4.29NASNMSALVEE58 pKa = 4.42GFTSQHH64 pKa = 5.92ANNTLFLSSGDD75 pKa = 3.48NYY77 pKa = 10.52IPGSRR82 pKa = 11.84YY83 pKa = 9.73DD84 pKa = 3.45AAADD88 pKa = 3.51EE89 pKa = 4.86SMVNVDD95 pKa = 4.07TIGVPGVARR104 pKa = 11.84ADD106 pKa = 3.25IAMLNAMGLQASAVGNHH123 pKa = 6.6DD124 pKa = 4.28LDD126 pKa = 4.5GGSAEE131 pKa = 4.2FASIINTQSASSCDD145 pKa = 3.72ANISCSADD153 pKa = 3.33YY154 pKa = 10.73IGAQFPYY161 pKa = 10.35LSANMDD167 pKa = 3.74FATDD171 pKa = 3.43EE172 pKa = 4.06NTIALVATAGQSAANLANKK191 pKa = 8.22LTASTVITVDD201 pKa = 3.43GEE203 pKa = 4.4HH204 pKa = 6.86IGIVGITTPTNDD216 pKa = 5.62DD217 pKa = 3.3ITTVDD222 pKa = 4.88DD223 pKa = 3.67IAVYY227 pKa = 9.76PLNDD231 pKa = 5.57DD232 pKa = 3.46IGEE235 pKa = 4.14LAAIVQSEE243 pKa = 4.57VNALTATGINKK254 pKa = 9.75IIALAHH260 pKa = 5.3MQSITYY266 pKa = 8.2EE267 pKa = 4.0KK268 pKa = 10.54QLAEE272 pKa = 4.39LLRR275 pKa = 11.84DD276 pKa = 3.15VDD278 pKa = 4.21IIVAGGSNTLLANDD292 pKa = 5.48DD293 pKa = 3.68DD294 pKa = 4.84RR295 pKa = 11.84LWAGDD300 pKa = 3.76TAAGSYY306 pKa = 10.01PIIKK310 pKa = 9.8QDD312 pKa = 3.48ADD314 pKa = 3.8GNDD317 pKa = 3.36VAIVNVDD324 pKa = 2.99ADD326 pKa = 3.83YY327 pKa = 10.97KK328 pKa = 9.88YY329 pKa = 10.82LGRR332 pKa = 11.84LVVDD336 pKa = 4.78FDD338 pKa = 4.92SDD340 pKa = 3.7GKK342 pKa = 11.26LLVDD346 pKa = 5.21SISAADD352 pKa = 3.96SGPVITDD359 pKa = 3.23GQMVDD364 pKa = 3.3ATEE367 pKa = 4.42GATPNSDD374 pKa = 3.35VIALVDD380 pKa = 4.33SINEE384 pKa = 3.74VLIEE388 pKa = 4.15SEE390 pKa = 4.69SNVVGHH396 pKa = 6.53TDD398 pKa = 2.87VFLNGTRR405 pKa = 11.84SYY407 pKa = 11.63VRR409 pKa = 11.84TEE411 pKa = 3.66EE412 pKa = 4.28TNLGNLSADD421 pKa = 3.27ANLWYY426 pKa = 10.34AQLHH430 pKa = 6.14DD431 pKa = 4.15ASVVISLKK439 pKa = 10.61NGGGIRR445 pKa = 11.84ADD447 pKa = 2.97IGYY450 pKa = 9.33SAFPAGSTDD459 pKa = 3.85PDD461 pKa = 3.99DD462 pKa = 3.72LTYY465 pKa = 10.65YY466 pKa = 10.22PPAAYY471 pKa = 8.25PAAQKK476 pKa = 11.19AEE478 pKa = 4.05GDD480 pKa = 3.08ISQYY484 pKa = 11.27DD485 pKa = 3.79VQTALAFNNGLSVFDD500 pKa = 3.94LTGEE504 pKa = 4.1QLWDD508 pKa = 3.3ILEE511 pKa = 4.38SGVAGVEE518 pKa = 4.14NISGGFHH525 pKa = 6.31HH526 pKa = 7.24VAGLCFSYY534 pKa = 10.9DD535 pKa = 3.18GTKK538 pKa = 9.21TARR541 pKa = 11.84QSDD544 pKa = 4.49PEE546 pKa = 4.4SGAVTVAGEE555 pKa = 4.21RR556 pKa = 11.84VQQIIVDD563 pKa = 3.8TDD565 pKa = 3.18SDD567 pKa = 4.54GNCDD571 pKa = 3.63KK572 pKa = 11.4NSDD575 pKa = 4.01DD576 pKa = 3.69MVMAGGTLQLPDD588 pKa = 3.09ATYY591 pKa = 8.74RR592 pKa = 11.84TVSLDD597 pKa = 3.47YY598 pKa = 10.34LASGAPYY605 pKa = 9.6PCAGDD610 pKa = 4.64DD611 pKa = 5.0CDD613 pKa = 4.58NQVQLGDD620 pKa = 4.02EE621 pKa = 4.36MTTDD625 pKa = 3.76PQASNFAATGSEE637 pKa = 3.54QDD639 pKa = 3.49ALAEE643 pKa = 3.98FLQAFYY649 pKa = 11.22ADD651 pKa = 4.16SDD653 pKa = 4.01SAYY656 pKa = 10.61NSSDD660 pKa = 3.46SVDD663 pKa = 3.36GGEE666 pKa = 3.78ADD668 pKa = 3.46ARR670 pKa = 11.84VIRR673 pKa = 11.84LDD675 pKa = 3.3KK676 pKa = 11.25
MM1 pKa = 7.19KK2 pKa = 10.31RR3 pKa = 11.84SLVTLGVIVALAGCDD18 pKa = 3.48SDD20 pKa = 3.54NHH22 pKa = 5.83NVVTSDD28 pKa = 3.34FTLQLLHH35 pKa = 6.33VADD38 pKa = 4.35MDD40 pKa = 4.1GSNATVLEE48 pKa = 4.29NASNMSALVEE58 pKa = 4.42GFTSQHH64 pKa = 5.92ANNTLFLSSGDD75 pKa = 3.48NYY77 pKa = 10.52IPGSRR82 pKa = 11.84YY83 pKa = 9.73DD84 pKa = 3.45AAADD88 pKa = 3.51EE89 pKa = 4.86SMVNVDD95 pKa = 4.07TIGVPGVARR104 pKa = 11.84ADD106 pKa = 3.25IAMLNAMGLQASAVGNHH123 pKa = 6.6DD124 pKa = 4.28LDD126 pKa = 4.5GGSAEE131 pKa = 4.2FASIINTQSASSCDD145 pKa = 3.72ANISCSADD153 pKa = 3.33YY154 pKa = 10.73IGAQFPYY161 pKa = 10.35LSANMDD167 pKa = 3.74FATDD171 pKa = 3.43EE172 pKa = 4.06NTIALVATAGQSAANLANKK191 pKa = 8.22LTASTVITVDD201 pKa = 3.43GEE203 pKa = 4.4HH204 pKa = 6.86IGIVGITTPTNDD216 pKa = 5.62DD217 pKa = 3.3ITTVDD222 pKa = 4.88DD223 pKa = 3.67IAVYY227 pKa = 9.76PLNDD231 pKa = 5.57DD232 pKa = 3.46IGEE235 pKa = 4.14LAAIVQSEE243 pKa = 4.57VNALTATGINKK254 pKa = 9.75IIALAHH260 pKa = 5.3MQSITYY266 pKa = 8.2EE267 pKa = 4.0KK268 pKa = 10.54QLAEE272 pKa = 4.39LLRR275 pKa = 11.84DD276 pKa = 3.15VDD278 pKa = 4.21IIVAGGSNTLLANDD292 pKa = 5.48DD293 pKa = 3.68DD294 pKa = 4.84RR295 pKa = 11.84LWAGDD300 pKa = 3.76TAAGSYY306 pKa = 10.01PIIKK310 pKa = 9.8QDD312 pKa = 3.48ADD314 pKa = 3.8GNDD317 pKa = 3.36VAIVNVDD324 pKa = 2.99ADD326 pKa = 3.83YY327 pKa = 10.97KK328 pKa = 9.88YY329 pKa = 10.82LGRR332 pKa = 11.84LVVDD336 pKa = 4.78FDD338 pKa = 4.92SDD340 pKa = 3.7GKK342 pKa = 11.26LLVDD346 pKa = 5.21SISAADD352 pKa = 3.96SGPVITDD359 pKa = 3.23GQMVDD364 pKa = 3.3ATEE367 pKa = 4.42GATPNSDD374 pKa = 3.35VIALVDD380 pKa = 4.33SINEE384 pKa = 3.74VLIEE388 pKa = 4.15SEE390 pKa = 4.69SNVVGHH396 pKa = 6.53TDD398 pKa = 2.87VFLNGTRR405 pKa = 11.84SYY407 pKa = 11.63VRR409 pKa = 11.84TEE411 pKa = 3.66EE412 pKa = 4.28TNLGNLSADD421 pKa = 3.27ANLWYY426 pKa = 10.34AQLHH430 pKa = 6.14DD431 pKa = 4.15ASVVISLKK439 pKa = 10.61NGGGIRR445 pKa = 11.84ADD447 pKa = 2.97IGYY450 pKa = 9.33SAFPAGSTDD459 pKa = 3.85PDD461 pKa = 3.99DD462 pKa = 3.72LTYY465 pKa = 10.65YY466 pKa = 10.22PPAAYY471 pKa = 8.25PAAQKK476 pKa = 11.19AEE478 pKa = 4.05GDD480 pKa = 3.08ISQYY484 pKa = 11.27DD485 pKa = 3.79VQTALAFNNGLSVFDD500 pKa = 3.94LTGEE504 pKa = 4.1QLWDD508 pKa = 3.3ILEE511 pKa = 4.38SGVAGVEE518 pKa = 4.14NISGGFHH525 pKa = 6.31HH526 pKa = 7.24VAGLCFSYY534 pKa = 10.9DD535 pKa = 3.18GTKK538 pKa = 9.21TARR541 pKa = 11.84QSDD544 pKa = 4.49PEE546 pKa = 4.4SGAVTVAGEE555 pKa = 4.21RR556 pKa = 11.84VQQIIVDD563 pKa = 3.8TDD565 pKa = 3.18SDD567 pKa = 4.54GNCDD571 pKa = 3.63KK572 pKa = 11.4NSDD575 pKa = 4.01DD576 pKa = 3.69MVMAGGTLQLPDD588 pKa = 3.09ATYY591 pKa = 8.74RR592 pKa = 11.84TVSLDD597 pKa = 3.47YY598 pKa = 10.34LASGAPYY605 pKa = 9.6PCAGDD610 pKa = 4.64DD611 pKa = 5.0CDD613 pKa = 4.58NQVQLGDD620 pKa = 4.02EE621 pKa = 4.36MTTDD625 pKa = 3.76PQASNFAATGSEE637 pKa = 3.54QDD639 pKa = 3.49ALAEE643 pKa = 3.98FLQAFYY649 pKa = 11.22ADD651 pKa = 4.16SDD653 pKa = 4.01SAYY656 pKa = 10.61NSSDD660 pKa = 3.46SVDD663 pKa = 3.36GGEE666 pKa = 3.78ADD668 pKa = 3.46ARR670 pKa = 11.84VIRR673 pKa = 11.84LDD675 pKa = 3.3KK676 pKa = 11.25
Molecular weight: 70.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6H1UGT7|A0A6H1UGT7_9GAMM GGDEF domain-containing protein OS=Ferrimonas lipolytica OX=2724191 GN=HER31_16195 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1156617 |
28 |
4188 |
326.8 |
36.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.778 ± 0.049 | 1.117 ± 0.016 |
5.637 ± 0.044 | 5.746 ± 0.035 |
3.719 ± 0.029 | 7.234 ± 0.04 |
2.32 ± 0.023 | 5.62 ± 0.032 |
4.224 ± 0.036 | 10.827 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.645 ± 0.021 | 3.965 ± 0.029 |
4.142 ± 0.027 | 5.464 ± 0.056 |
4.915 ± 0.036 | 6.252 ± 0.038 |
5.254 ± 0.042 | 7.022 ± 0.031 |
1.309 ± 0.019 | 2.809 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
