
Capybara microvirus Cap3_SP_374
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
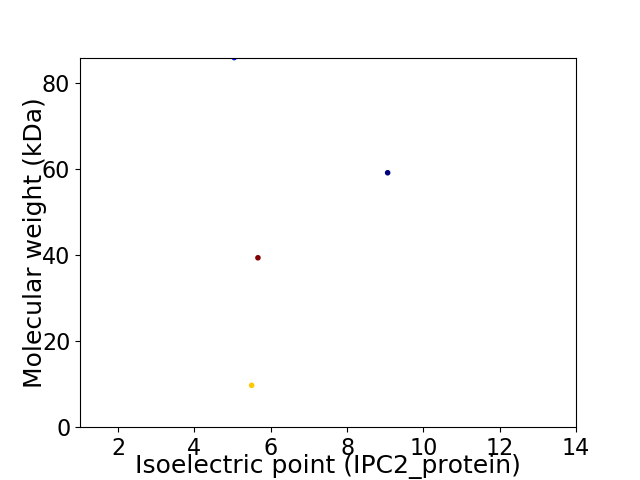
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8F7|A0A4P8W8F7_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_374 OX=2585437 PE=3 SV=1
MM1 pKa = 7.39SKK3 pKa = 10.66NVFIEE8 pKa = 4.27RR9 pKa = 11.84NDD11 pKa = 3.51FEE13 pKa = 6.21NDD15 pKa = 3.12VKK17 pKa = 10.99RR18 pKa = 11.84SAFDD22 pKa = 3.84LSHH25 pKa = 6.73NNHH28 pKa = 6.7LSLKK32 pKa = 9.33LGKK35 pKa = 8.4ITPVFLQEE43 pKa = 4.31GLPGSTFKK51 pKa = 10.44IDD53 pKa = 3.21PVMALKK59 pKa = 10.19AAPLVFPMQTKK70 pKa = 10.41LKK72 pKa = 10.49ASLSFFKK79 pKa = 11.28VNTRR83 pKa = 11.84TLWKK87 pKa = 10.11DD88 pKa = 2.66WTKK91 pKa = 10.82FITNVEE97 pKa = 4.31HH98 pKa = 7.63DD99 pKa = 3.45STMPKK104 pKa = 10.12VSVSADD110 pKa = 3.3SNFWKK115 pKa = 9.46TGEE118 pKa = 4.04LADD121 pKa = 3.86YY122 pKa = 10.78LGIPTAIYY130 pKa = 10.34SSSEE134 pKa = 3.62LTDD137 pKa = 3.2YY138 pKa = 8.72RR139 pKa = 11.84TLHH142 pKa = 6.77ALTEE146 pKa = 4.2VQDD149 pKa = 5.01LIKK152 pKa = 10.51GWYY155 pKa = 8.63LRR157 pKa = 11.84YY158 pKa = 9.99RR159 pKa = 11.84EE160 pKa = 4.85DD161 pKa = 4.67DD162 pKa = 3.02ASYY165 pKa = 10.51WMKK168 pKa = 11.11APINTTPSMNMKK180 pKa = 10.18GYY182 pKa = 10.15IGVAPSIDD190 pKa = 3.61QTRR193 pKa = 11.84SLYY196 pKa = 9.99EE197 pKa = 3.67QILVGRR203 pKa = 11.84NDD205 pKa = 3.7MVQQPYY211 pKa = 10.51GFPAMSLTGYY221 pKa = 9.97DD222 pKa = 3.89GPGSSQQLTKK232 pKa = 10.62SFPRR236 pKa = 11.84ISRR239 pKa = 11.84VFAITEE245 pKa = 3.87QVKK248 pKa = 10.58IGDD251 pKa = 3.96VTCSFNSNTNTAVINFVRR269 pKa = 11.84ADD271 pKa = 3.42GTMLFYY277 pKa = 10.42YY278 pKa = 9.88TGTITAHH285 pKa = 7.03KK286 pKa = 9.28YY287 pKa = 10.61SCTLTQANVNVINNEE302 pKa = 3.71IANGSPVFVFAYY314 pKa = 8.86TNQNFTTEE322 pKa = 4.59ASFQPDD328 pKa = 3.16TVEE331 pKa = 4.44YY332 pKa = 10.12IGEE335 pKa = 4.3TLTVPVALTYY345 pKa = 10.61TKK347 pKa = 10.85DD348 pKa = 3.33FTDD351 pKa = 3.04ISEE354 pKa = 4.4DD355 pKa = 3.49PEE357 pKa = 4.19RR358 pKa = 11.84NPFANGSTQRR368 pKa = 11.84NVVPISALPFRR379 pKa = 11.84AYY381 pKa = 9.7EE382 pKa = 3.96AVYY385 pKa = 9.5NAYY388 pKa = 9.96YY389 pKa = 10.36RR390 pKa = 11.84NEE392 pKa = 4.11RR393 pKa = 11.84VDD395 pKa = 3.57PLIINGRR402 pKa = 11.84KK403 pKa = 9.68EE404 pKa = 3.52YY405 pKa = 10.8DD406 pKa = 3.43LYY408 pKa = 11.37CNQEE412 pKa = 3.26GGYY415 pKa = 10.7DD416 pKa = 3.27HH417 pKa = 7.38TEE419 pKa = 4.12YY420 pKa = 11.03LLHH423 pKa = 6.86DD424 pKa = 5.13APWEE428 pKa = 4.03TDD430 pKa = 3.18VYY432 pKa = 9.55TSCLPSPMDD441 pKa = 4.06GPAPLIGLQSNLIGDD456 pKa = 5.14KK457 pKa = 9.49ITATMTTEE465 pKa = 3.54SGEE468 pKa = 4.14THH470 pKa = 7.28DD471 pKa = 4.35YY472 pKa = 11.09EE473 pKa = 5.63IGAVTDD479 pKa = 3.4STGNITGFTYY489 pKa = 10.64SEE491 pKa = 5.28DD492 pKa = 3.2IPYY495 pKa = 10.72DD496 pKa = 3.41LQRR499 pKa = 11.84QLNQKK504 pKa = 8.47VLSGISVNTIRR515 pKa = 11.84NASAFEE521 pKa = 3.99RR522 pKa = 11.84YY523 pKa = 8.97LRR525 pKa = 11.84NGLRR529 pKa = 11.84RR530 pKa = 11.84GKK532 pKa = 10.51KK533 pKa = 10.31YY534 pKa = 9.79IDD536 pKa = 3.22NMKK539 pKa = 10.29SHH541 pKa = 7.49FGDD544 pKa = 3.12THH546 pKa = 6.59IQYY549 pKa = 10.98DD550 pKa = 4.35SLQMPTFHH558 pKa = 7.15GGITSVFNVNQISSTTEE575 pKa = 3.34NPAIDD580 pKa = 4.22LALGSYY586 pKa = 9.48AGQMSLIDD594 pKa = 4.28NPNHH598 pKa = 6.54TISIYY603 pKa = 10.65CEE605 pKa = 3.68EE606 pKa = 4.03HH607 pKa = 6.85CYY609 pKa = 10.36IIGLLTVTPVPIYY622 pKa = 10.77NQTLPKK628 pKa = 10.38LFSKK632 pKa = 10.53FDD634 pKa = 3.27KK635 pKa = 10.76FDD637 pKa = 4.15FYY639 pKa = 11.23TPEE642 pKa = 4.06FDD644 pKa = 3.74QLSMQPIKK652 pKa = 10.4RR653 pKa = 11.84QEE655 pKa = 3.81VSVTNTYY662 pKa = 11.38AEE664 pKa = 4.37GGYY667 pKa = 6.66TACNEE672 pKa = 4.1TFGYY676 pKa = 8.96QLPWYY681 pKa = 8.45EE682 pKa = 4.29YY683 pKa = 11.19KK684 pKa = 10.72NALDD688 pKa = 3.75TTHH691 pKa = 6.31GQMRR695 pKa = 11.84TTMKK699 pKa = 10.11DD700 pKa = 3.26YY701 pKa = 11.04NMNRR705 pKa = 11.84VFHH708 pKa = 7.04GIPQLSSDD716 pKa = 4.92FIYY719 pKa = 10.96CDD721 pKa = 3.73GEE723 pKa = 4.14TLNNVFAVADD733 pKa = 3.79DD734 pKa = 4.37TEE736 pKa = 5.0DD737 pKa = 3.35KK738 pKa = 11.38VFGQIYY744 pKa = 9.95FKK746 pKa = 8.93TTVAHH751 pKa = 5.52PMSRR755 pKa = 11.84IGEE758 pKa = 4.48PTWHH762 pKa = 6.71
MM1 pKa = 7.39SKK3 pKa = 10.66NVFIEE8 pKa = 4.27RR9 pKa = 11.84NDD11 pKa = 3.51FEE13 pKa = 6.21NDD15 pKa = 3.12VKK17 pKa = 10.99RR18 pKa = 11.84SAFDD22 pKa = 3.84LSHH25 pKa = 6.73NNHH28 pKa = 6.7LSLKK32 pKa = 9.33LGKK35 pKa = 8.4ITPVFLQEE43 pKa = 4.31GLPGSTFKK51 pKa = 10.44IDD53 pKa = 3.21PVMALKK59 pKa = 10.19AAPLVFPMQTKK70 pKa = 10.41LKK72 pKa = 10.49ASLSFFKK79 pKa = 11.28VNTRR83 pKa = 11.84TLWKK87 pKa = 10.11DD88 pKa = 2.66WTKK91 pKa = 10.82FITNVEE97 pKa = 4.31HH98 pKa = 7.63DD99 pKa = 3.45STMPKK104 pKa = 10.12VSVSADD110 pKa = 3.3SNFWKK115 pKa = 9.46TGEE118 pKa = 4.04LADD121 pKa = 3.86YY122 pKa = 10.78LGIPTAIYY130 pKa = 10.34SSSEE134 pKa = 3.62LTDD137 pKa = 3.2YY138 pKa = 8.72RR139 pKa = 11.84TLHH142 pKa = 6.77ALTEE146 pKa = 4.2VQDD149 pKa = 5.01LIKK152 pKa = 10.51GWYY155 pKa = 8.63LRR157 pKa = 11.84YY158 pKa = 9.99RR159 pKa = 11.84EE160 pKa = 4.85DD161 pKa = 4.67DD162 pKa = 3.02ASYY165 pKa = 10.51WMKK168 pKa = 11.11APINTTPSMNMKK180 pKa = 10.18GYY182 pKa = 10.15IGVAPSIDD190 pKa = 3.61QTRR193 pKa = 11.84SLYY196 pKa = 9.99EE197 pKa = 3.67QILVGRR203 pKa = 11.84NDD205 pKa = 3.7MVQQPYY211 pKa = 10.51GFPAMSLTGYY221 pKa = 9.97DD222 pKa = 3.89GPGSSQQLTKK232 pKa = 10.62SFPRR236 pKa = 11.84ISRR239 pKa = 11.84VFAITEE245 pKa = 3.87QVKK248 pKa = 10.58IGDD251 pKa = 3.96VTCSFNSNTNTAVINFVRR269 pKa = 11.84ADD271 pKa = 3.42GTMLFYY277 pKa = 10.42YY278 pKa = 9.88TGTITAHH285 pKa = 7.03KK286 pKa = 9.28YY287 pKa = 10.61SCTLTQANVNVINNEE302 pKa = 3.71IANGSPVFVFAYY314 pKa = 8.86TNQNFTTEE322 pKa = 4.59ASFQPDD328 pKa = 3.16TVEE331 pKa = 4.44YY332 pKa = 10.12IGEE335 pKa = 4.3TLTVPVALTYY345 pKa = 10.61TKK347 pKa = 10.85DD348 pKa = 3.33FTDD351 pKa = 3.04ISEE354 pKa = 4.4DD355 pKa = 3.49PEE357 pKa = 4.19RR358 pKa = 11.84NPFANGSTQRR368 pKa = 11.84NVVPISALPFRR379 pKa = 11.84AYY381 pKa = 9.7EE382 pKa = 3.96AVYY385 pKa = 9.5NAYY388 pKa = 9.96YY389 pKa = 10.36RR390 pKa = 11.84NEE392 pKa = 4.11RR393 pKa = 11.84VDD395 pKa = 3.57PLIINGRR402 pKa = 11.84KK403 pKa = 9.68EE404 pKa = 3.52YY405 pKa = 10.8DD406 pKa = 3.43LYY408 pKa = 11.37CNQEE412 pKa = 3.26GGYY415 pKa = 10.7DD416 pKa = 3.27HH417 pKa = 7.38TEE419 pKa = 4.12YY420 pKa = 11.03LLHH423 pKa = 6.86DD424 pKa = 5.13APWEE428 pKa = 4.03TDD430 pKa = 3.18VYY432 pKa = 9.55TSCLPSPMDD441 pKa = 4.06GPAPLIGLQSNLIGDD456 pKa = 5.14KK457 pKa = 9.49ITATMTTEE465 pKa = 3.54SGEE468 pKa = 4.14THH470 pKa = 7.28DD471 pKa = 4.35YY472 pKa = 11.09EE473 pKa = 5.63IGAVTDD479 pKa = 3.4STGNITGFTYY489 pKa = 10.64SEE491 pKa = 5.28DD492 pKa = 3.2IPYY495 pKa = 10.72DD496 pKa = 3.41LQRR499 pKa = 11.84QLNQKK504 pKa = 8.47VLSGISVNTIRR515 pKa = 11.84NASAFEE521 pKa = 3.99RR522 pKa = 11.84YY523 pKa = 8.97LRR525 pKa = 11.84NGLRR529 pKa = 11.84RR530 pKa = 11.84GKK532 pKa = 10.51KK533 pKa = 10.31YY534 pKa = 9.79IDD536 pKa = 3.22NMKK539 pKa = 10.29SHH541 pKa = 7.49FGDD544 pKa = 3.12THH546 pKa = 6.59IQYY549 pKa = 10.98DD550 pKa = 4.35SLQMPTFHH558 pKa = 7.15GGITSVFNVNQISSTTEE575 pKa = 3.34NPAIDD580 pKa = 4.22LALGSYY586 pKa = 9.48AGQMSLIDD594 pKa = 4.28NPNHH598 pKa = 6.54TISIYY603 pKa = 10.65CEE605 pKa = 3.68EE606 pKa = 4.03HH607 pKa = 6.85CYY609 pKa = 10.36IIGLLTVTPVPIYY622 pKa = 10.77NQTLPKK628 pKa = 10.38LFSKK632 pKa = 10.53FDD634 pKa = 3.27KK635 pKa = 10.76FDD637 pKa = 4.15FYY639 pKa = 11.23TPEE642 pKa = 4.06FDD644 pKa = 3.74QLSMQPIKK652 pKa = 10.4RR653 pKa = 11.84QEE655 pKa = 3.81VSVTNTYY662 pKa = 11.38AEE664 pKa = 4.37GGYY667 pKa = 6.66TACNEE672 pKa = 4.1TFGYY676 pKa = 8.96QLPWYY681 pKa = 8.45EE682 pKa = 4.29YY683 pKa = 11.19KK684 pKa = 10.72NALDD688 pKa = 3.75TTHH691 pKa = 6.31GQMRR695 pKa = 11.84TTMKK699 pKa = 10.11DD700 pKa = 3.26YY701 pKa = 11.04NMNRR705 pKa = 11.84VFHH708 pKa = 7.04GIPQLSSDD716 pKa = 4.92FIYY719 pKa = 10.96CDD721 pKa = 3.73GEE723 pKa = 4.14TLNNVFAVADD733 pKa = 3.79DD734 pKa = 4.37TEE736 pKa = 5.0DD737 pKa = 3.35KK738 pKa = 11.38VFGQIYY744 pKa = 9.95FKK746 pKa = 8.93TTVAHH751 pKa = 5.52PMSRR755 pKa = 11.84IGEE758 pKa = 4.48PTWHH762 pKa = 6.71
Molecular weight: 86.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8E6|A0A4P8W8E6_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_374 OX=2585437 PE=4 SV=1
MM1 pKa = 7.57EE2 pKa = 5.59CLNPVTLIIEE12 pKa = 4.27KK13 pKa = 9.27TVYY16 pKa = 10.49NYY18 pKa = 9.05EE19 pKa = 4.01KK20 pKa = 9.91QIKK23 pKa = 8.86YY24 pKa = 10.72AVIDD28 pKa = 3.57NQRR31 pKa = 11.84YY32 pKa = 9.78DD33 pKa = 3.37GLSAYY38 pKa = 10.18DD39 pKa = 3.33IIFYY43 pKa = 10.12SRR45 pKa = 11.84KK46 pKa = 9.78ALEE49 pKa = 4.21NRR51 pKa = 11.84QSFPRR56 pKa = 11.84DD57 pKa = 3.16EE58 pKa = 4.17QNRR61 pKa = 11.84ILRR64 pKa = 11.84YY65 pKa = 9.66NPRR68 pKa = 11.84EE69 pKa = 3.64HH70 pKa = 6.13QFYY73 pKa = 10.48IRR75 pKa = 11.84VEE77 pKa = 4.01GVIDD81 pKa = 3.61GRR83 pKa = 11.84YY84 pKa = 7.39KK85 pKa = 10.38QKK87 pKa = 10.81SIYY90 pKa = 8.92FTQQVACRR98 pKa = 11.84HH99 pKa = 5.69CVLCVEE105 pKa = 5.12RR106 pKa = 11.84QRR108 pKa = 11.84QDD110 pKa = 2.31ITNRR114 pKa = 11.84ILFEE118 pKa = 4.25TEE120 pKa = 3.91TYY122 pKa = 10.0GPPIISTLTYY132 pKa = 10.66NNQSLPTDD140 pKa = 3.72GLNYY144 pKa = 10.46SDD146 pKa = 3.63VQKK149 pKa = 10.7LFKK152 pKa = 10.59VLRR155 pKa = 11.84QTITRR160 pKa = 11.84HH161 pKa = 4.72SAIFTDD167 pKa = 4.95GSGFRR172 pKa = 11.84YY173 pKa = 9.37FVAGEE178 pKa = 4.1YY179 pKa = 11.16GSITHH184 pKa = 7.05RR185 pKa = 11.84AHH187 pKa = 5.89YY188 pKa = 10.12HH189 pKa = 5.2LLLWNFPTPQQCKK202 pKa = 6.59EE203 pKa = 3.94WYY205 pKa = 8.32LKK207 pKa = 10.53KK208 pKa = 10.98YY209 pKa = 8.05NTEE212 pKa = 3.56LRR214 pKa = 11.84VAYY217 pKa = 10.05DD218 pKa = 3.53PNPNNKK224 pKa = 8.29NWYY227 pKa = 9.17FPHH230 pKa = 7.04LNNEE234 pKa = 4.13YY235 pKa = 10.26IIAEE239 pKa = 4.19KK240 pKa = 10.22KK241 pKa = 10.4GRR243 pKa = 11.84TFPVIEE249 pKa = 4.34STSYY253 pKa = 10.9LYY255 pKa = 10.1QDD257 pKa = 3.38YY258 pKa = 10.35FIRR261 pKa = 11.84YY262 pKa = 6.22HH263 pKa = 6.04WKK265 pKa = 9.99RR266 pKa = 11.84GLTLTEE272 pKa = 4.1SMRR275 pKa = 11.84GANTQSAATYY285 pKa = 8.3CAKK288 pKa = 10.39YY289 pKa = 9.8VNKK292 pKa = 10.28SASPHH297 pKa = 6.44PNPNMNPTFYY307 pKa = 10.58RR308 pKa = 11.84SSTGNGGLGSRR319 pKa = 11.84YY320 pKa = 7.66IRR322 pKa = 11.84EE323 pKa = 4.07YY324 pKa = 9.5IRR326 pKa = 11.84PYY328 pKa = 10.64ILTQYY333 pKa = 9.93HH334 pKa = 5.72ITGQVPLEE342 pKa = 4.02YY343 pKa = 10.31VYY345 pKa = 10.93RR346 pKa = 11.84PTNTTQIIHH355 pKa = 5.18VPIRR359 pKa = 11.84GYY361 pKa = 11.26LLDD364 pKa = 3.4QCFPTVSQLLPARR377 pKa = 11.84YY378 pKa = 8.44IEE380 pKa = 4.32YY381 pKa = 8.84YY382 pKa = 7.71QTVIARR388 pKa = 11.84SDD390 pKa = 3.18TLQRR394 pKa = 11.84LKK396 pKa = 11.26VKK398 pKa = 10.59DD399 pKa = 3.72LAPNNIIRR407 pKa = 11.84PMNTPRR413 pKa = 11.84GNNLNEE419 pKa = 3.64KK420 pKa = 9.49EE421 pKa = 3.98YY422 pKa = 11.02RR423 pKa = 11.84VFKK426 pKa = 10.76RR427 pKa = 11.84KK428 pKa = 9.79LSLPEE433 pKa = 3.91YY434 pKa = 10.8NEE436 pKa = 3.98MIQYY440 pKa = 10.87LKK442 pKa = 10.92DD443 pKa = 3.23KK444 pKa = 11.25SKK446 pKa = 11.19DD447 pKa = 3.07IDD449 pKa = 3.86FKK451 pKa = 11.37QVANMLQLRR460 pKa = 11.84KK461 pKa = 9.66IYY463 pKa = 10.4LEE465 pKa = 3.98NVAQQEE471 pKa = 4.16LPYY474 pKa = 10.44RR475 pKa = 11.84DD476 pKa = 4.3DD477 pKa = 5.83LSDD480 pKa = 4.04KK481 pKa = 10.43SIKK484 pKa = 9.59YY485 pKa = 8.71EE486 pKa = 3.7KK487 pKa = 10.43LINFRR492 pKa = 11.84KK493 pKa = 10.09NRR495 pKa = 11.84DD496 pKa = 3.48KK497 pKa = 11.1II498 pKa = 3.8
MM1 pKa = 7.57EE2 pKa = 5.59CLNPVTLIIEE12 pKa = 4.27KK13 pKa = 9.27TVYY16 pKa = 10.49NYY18 pKa = 9.05EE19 pKa = 4.01KK20 pKa = 9.91QIKK23 pKa = 8.86YY24 pKa = 10.72AVIDD28 pKa = 3.57NQRR31 pKa = 11.84YY32 pKa = 9.78DD33 pKa = 3.37GLSAYY38 pKa = 10.18DD39 pKa = 3.33IIFYY43 pKa = 10.12SRR45 pKa = 11.84KK46 pKa = 9.78ALEE49 pKa = 4.21NRR51 pKa = 11.84QSFPRR56 pKa = 11.84DD57 pKa = 3.16EE58 pKa = 4.17QNRR61 pKa = 11.84ILRR64 pKa = 11.84YY65 pKa = 9.66NPRR68 pKa = 11.84EE69 pKa = 3.64HH70 pKa = 6.13QFYY73 pKa = 10.48IRR75 pKa = 11.84VEE77 pKa = 4.01GVIDD81 pKa = 3.61GRR83 pKa = 11.84YY84 pKa = 7.39KK85 pKa = 10.38QKK87 pKa = 10.81SIYY90 pKa = 8.92FTQQVACRR98 pKa = 11.84HH99 pKa = 5.69CVLCVEE105 pKa = 5.12RR106 pKa = 11.84QRR108 pKa = 11.84QDD110 pKa = 2.31ITNRR114 pKa = 11.84ILFEE118 pKa = 4.25TEE120 pKa = 3.91TYY122 pKa = 10.0GPPIISTLTYY132 pKa = 10.66NNQSLPTDD140 pKa = 3.72GLNYY144 pKa = 10.46SDD146 pKa = 3.63VQKK149 pKa = 10.7LFKK152 pKa = 10.59VLRR155 pKa = 11.84QTITRR160 pKa = 11.84HH161 pKa = 4.72SAIFTDD167 pKa = 4.95GSGFRR172 pKa = 11.84YY173 pKa = 9.37FVAGEE178 pKa = 4.1YY179 pKa = 11.16GSITHH184 pKa = 7.05RR185 pKa = 11.84AHH187 pKa = 5.89YY188 pKa = 10.12HH189 pKa = 5.2LLLWNFPTPQQCKK202 pKa = 6.59EE203 pKa = 3.94WYY205 pKa = 8.32LKK207 pKa = 10.53KK208 pKa = 10.98YY209 pKa = 8.05NTEE212 pKa = 3.56LRR214 pKa = 11.84VAYY217 pKa = 10.05DD218 pKa = 3.53PNPNNKK224 pKa = 8.29NWYY227 pKa = 9.17FPHH230 pKa = 7.04LNNEE234 pKa = 4.13YY235 pKa = 10.26IIAEE239 pKa = 4.19KK240 pKa = 10.22KK241 pKa = 10.4GRR243 pKa = 11.84TFPVIEE249 pKa = 4.34STSYY253 pKa = 10.9LYY255 pKa = 10.1QDD257 pKa = 3.38YY258 pKa = 10.35FIRR261 pKa = 11.84YY262 pKa = 6.22HH263 pKa = 6.04WKK265 pKa = 9.99RR266 pKa = 11.84GLTLTEE272 pKa = 4.1SMRR275 pKa = 11.84GANTQSAATYY285 pKa = 8.3CAKK288 pKa = 10.39YY289 pKa = 9.8VNKK292 pKa = 10.28SASPHH297 pKa = 6.44PNPNMNPTFYY307 pKa = 10.58RR308 pKa = 11.84SSTGNGGLGSRR319 pKa = 11.84YY320 pKa = 7.66IRR322 pKa = 11.84EE323 pKa = 4.07YY324 pKa = 9.5IRR326 pKa = 11.84PYY328 pKa = 10.64ILTQYY333 pKa = 9.93HH334 pKa = 5.72ITGQVPLEE342 pKa = 4.02YY343 pKa = 10.31VYY345 pKa = 10.93RR346 pKa = 11.84PTNTTQIIHH355 pKa = 5.18VPIRR359 pKa = 11.84GYY361 pKa = 11.26LLDD364 pKa = 3.4QCFPTVSQLLPARR377 pKa = 11.84YY378 pKa = 8.44IEE380 pKa = 4.32YY381 pKa = 8.84YY382 pKa = 7.71QTVIARR388 pKa = 11.84SDD390 pKa = 3.18TLQRR394 pKa = 11.84LKK396 pKa = 11.26VKK398 pKa = 10.59DD399 pKa = 3.72LAPNNIIRR407 pKa = 11.84PMNTPRR413 pKa = 11.84GNNLNEE419 pKa = 3.64KK420 pKa = 9.49EE421 pKa = 3.98YY422 pKa = 11.02RR423 pKa = 11.84VFKK426 pKa = 10.76RR427 pKa = 11.84KK428 pKa = 9.79LSLPEE433 pKa = 3.91YY434 pKa = 10.8NEE436 pKa = 3.98MIQYY440 pKa = 10.87LKK442 pKa = 10.92DD443 pKa = 3.23KK444 pKa = 11.25SKK446 pKa = 11.19DD447 pKa = 3.07IDD449 pKa = 3.86FKK451 pKa = 11.37QVANMLQLRR460 pKa = 11.84KK461 pKa = 9.66IYY463 pKa = 10.4LEE465 pKa = 3.98NVAQQEE471 pKa = 4.16LPYY474 pKa = 10.44RR475 pKa = 11.84DD476 pKa = 4.3DD477 pKa = 5.83LSDD480 pKa = 4.04KK481 pKa = 10.43SIKK484 pKa = 9.59YY485 pKa = 8.71EE486 pKa = 3.7KK487 pKa = 10.43LINFRR492 pKa = 11.84KK493 pKa = 10.09NRR495 pKa = 11.84DD496 pKa = 3.48KK497 pKa = 11.1II498 pKa = 3.8
Molecular weight: 59.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1697 |
88 |
762 |
424.3 |
48.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.246 ± 1.458 | 0.943 ± 0.24 |
5.48 ± 0.524 | 5.716 ± 0.589 |
3.654 ± 0.986 | 5.127 ± 0.653 |
1.768 ± 0.419 | 6.364 ± 0.516 |
5.716 ± 0.711 | 7.778 ± 0.467 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.298 ± 0.432 | 7.307 ± 0.6 |
4.655 ± 0.728 | 6.011 ± 1.294 |
5.186 ± 1.143 | 6.011 ± 0.642 |
7.778 ± 1.153 | 4.891 ± 0.526 |
0.884 ± 0.108 | 6.187 ± 1.073 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
