
Momordica charantia (Bitter gourd) (Balsam pear)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Momordiceae; Momordica
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
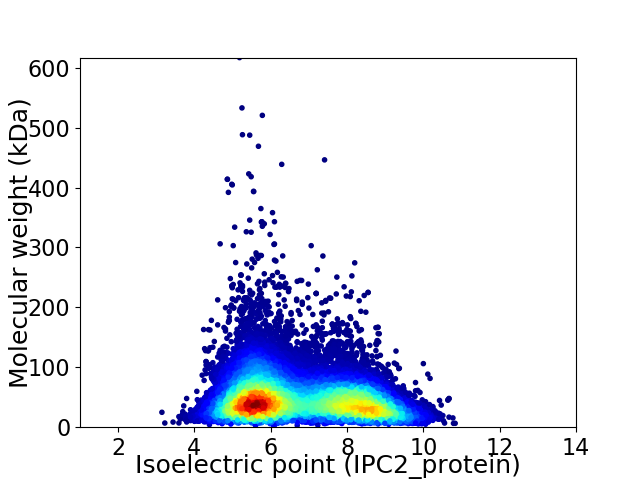
Virtual 2D-PAGE plot for 24040 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
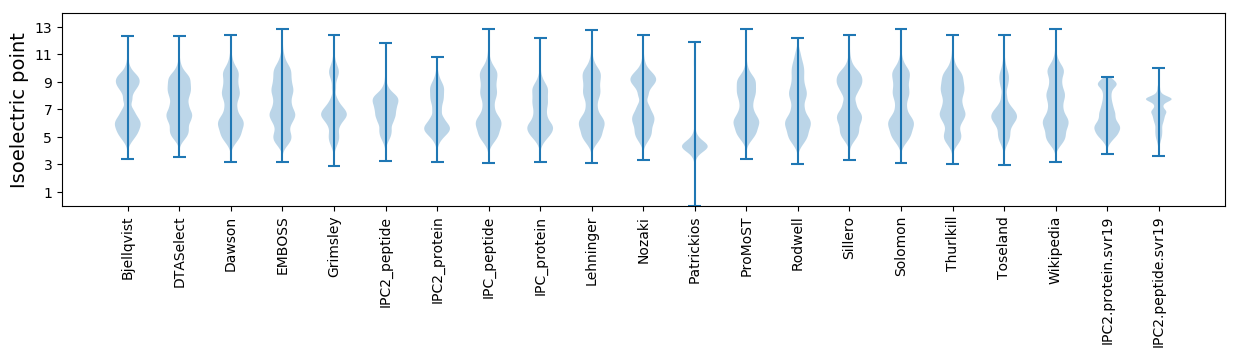
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6J1DIT2|A0A6J1DIT2_MOMCH Lipase OS=Momordica charantia OX=3673 GN=LOC111021471 PE=3 SV=1
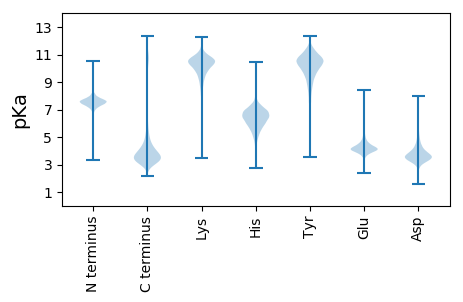
MM1 pKa = 7.61ALASVEE7 pKa = 4.16GRR9 pKa = 11.84STLNPNAPLFIPAAYY24 pKa = 9.38QVEE27 pKa = 4.54DD28 pKa = 5.92FSPQWWQLVTTSTWYY43 pKa = 9.91RR44 pKa = 11.84DD45 pKa = 3.4YY46 pKa = 10.77WLSQHH51 pKa = 6.04QEE53 pKa = 3.55EE54 pKa = 4.56SDD56 pKa = 4.14FYY58 pKa = 11.17IEE60 pKa = 6.48DD61 pKa = 4.23DD62 pKa = 4.25FNSNDD67 pKa = 3.46IADD70 pKa = 5.14LLPEE74 pKa = 5.15AFDD77 pKa = 4.31LDD79 pKa = 4.03ANEE82 pKa = 4.68EE83 pKa = 4.19LRR85 pKa = 11.84TMEE88 pKa = 4.62AEE90 pKa = 3.88FEE92 pKa = 4.21EE93 pKa = 5.53FIQASLTEE101 pKa = 4.05AHH103 pKa = 6.58HH104 pKa = 7.1PEE106 pKa = 4.0MM107 pKa = 5.79
MM1 pKa = 7.61ALASVEE7 pKa = 4.16GRR9 pKa = 11.84STLNPNAPLFIPAAYY24 pKa = 9.38QVEE27 pKa = 4.54DD28 pKa = 5.92FSPQWWQLVTTSTWYY43 pKa = 9.91RR44 pKa = 11.84DD45 pKa = 3.4YY46 pKa = 10.77WLSQHH51 pKa = 6.04QEE53 pKa = 3.55EE54 pKa = 4.56SDD56 pKa = 4.14FYY58 pKa = 11.17IEE60 pKa = 6.48DD61 pKa = 4.23DD62 pKa = 4.25FNSNDD67 pKa = 3.46IADD70 pKa = 5.14LLPEE74 pKa = 5.15AFDD77 pKa = 4.31LDD79 pKa = 4.03ANEE82 pKa = 4.68EE83 pKa = 4.19LRR85 pKa = 11.84TMEE88 pKa = 4.62AEE90 pKa = 3.88FEE92 pKa = 4.21EE93 pKa = 5.53FIQASLTEE101 pKa = 4.05AHH103 pKa = 6.58HH104 pKa = 7.1PEE106 pKa = 4.0MM107 pKa = 5.79
Molecular weight: 12.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J1DPD2|A0A6J1DPD2_MOMCH uncharacterized protein LOC111023073 isoform X1 OS=Momordica charantia OX=3673 GN=LOC111023073 PE=4 SV=1
MM1 pKa = 7.81PNLPDD6 pKa = 3.06IMGNVASSVASGFFSAVGKK25 pKa = 9.77LFRR28 pKa = 11.84SPLDD32 pKa = 3.5FLSGKK37 pKa = 9.74SCSSVCGSTWDD48 pKa = 4.39FICYY52 pKa = 9.68IEE54 pKa = 4.35NFCVANLLKK63 pKa = 10.8LGMVLILSLFVILLLYY79 pKa = 10.19LLHH82 pKa = 7.15KK83 pKa = 10.38IGIFGCICRR92 pKa = 11.84GLCRR96 pKa = 11.84MTWTCIASYY105 pKa = 9.68FYY107 pKa = 11.19AWDD110 pKa = 3.7YY111 pKa = 11.29CCTFMCIKK119 pKa = 10.15LGSVKK124 pKa = 9.02RR125 pKa = 11.84TRR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84HH131 pKa = 4.99RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84DD135 pKa = 3.35LEE137 pKa = 4.2EE138 pKa = 4.02EE139 pKa = 4.27FEE141 pKa = 4.58SEE143 pKa = 4.26GGKK146 pKa = 9.29HH147 pKa = 6.01RR148 pKa = 11.84YY149 pKa = 9.2GSSSDD154 pKa = 3.33SSSVPEE160 pKa = 4.39RR161 pKa = 11.84IEE163 pKa = 3.94LRR165 pKa = 11.84SSQRR169 pKa = 11.84ASRR172 pKa = 11.84RR173 pKa = 11.84WRR175 pKa = 11.84MNHH178 pKa = 6.8RR179 pKa = 11.84GSQMRR184 pKa = 11.84KK185 pKa = 8.57ALRR188 pKa = 11.84PKK190 pKa = 10.26SRR192 pKa = 11.84GIRR195 pKa = 11.84VRR197 pKa = 11.84SGRR200 pKa = 11.84TLVYY204 pKa = 9.95GKK206 pKa = 9.61HH207 pKa = 5.27RR208 pKa = 11.84RR209 pKa = 11.84KK210 pKa = 10.27SSEE213 pKa = 3.71VVNRR217 pKa = 11.84LGEE220 pKa = 3.91IHH222 pKa = 6.71SLGRR226 pKa = 11.84HH227 pKa = 4.9GSSKK231 pKa = 10.07FVHH234 pKa = 6.09EE235 pKa = 4.69EE236 pKa = 3.06IRR238 pKa = 11.84YY239 pKa = 9.21KK240 pKa = 10.57RR241 pKa = 11.84GRR243 pKa = 11.84QKK245 pKa = 11.28
MM1 pKa = 7.81PNLPDD6 pKa = 3.06IMGNVASSVASGFFSAVGKK25 pKa = 9.77LFRR28 pKa = 11.84SPLDD32 pKa = 3.5FLSGKK37 pKa = 9.74SCSSVCGSTWDD48 pKa = 4.39FICYY52 pKa = 9.68IEE54 pKa = 4.35NFCVANLLKK63 pKa = 10.8LGMVLILSLFVILLLYY79 pKa = 10.19LLHH82 pKa = 7.15KK83 pKa = 10.38IGIFGCICRR92 pKa = 11.84GLCRR96 pKa = 11.84MTWTCIASYY105 pKa = 9.68FYY107 pKa = 11.19AWDD110 pKa = 3.7YY111 pKa = 11.29CCTFMCIKK119 pKa = 10.15LGSVKK124 pKa = 9.02RR125 pKa = 11.84TRR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84HH131 pKa = 4.99RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84DD135 pKa = 3.35LEE137 pKa = 4.2EE138 pKa = 4.02EE139 pKa = 4.27FEE141 pKa = 4.58SEE143 pKa = 4.26GGKK146 pKa = 9.29HH147 pKa = 6.01RR148 pKa = 11.84YY149 pKa = 9.2GSSSDD154 pKa = 3.33SSSVPEE160 pKa = 4.39RR161 pKa = 11.84IEE163 pKa = 3.94LRR165 pKa = 11.84SSQRR169 pKa = 11.84ASRR172 pKa = 11.84RR173 pKa = 11.84WRR175 pKa = 11.84MNHH178 pKa = 6.8RR179 pKa = 11.84GSQMRR184 pKa = 11.84KK185 pKa = 8.57ALRR188 pKa = 11.84PKK190 pKa = 10.26SRR192 pKa = 11.84GIRR195 pKa = 11.84VRR197 pKa = 11.84SGRR200 pKa = 11.84TLVYY204 pKa = 9.95GKK206 pKa = 9.61HH207 pKa = 5.27RR208 pKa = 11.84RR209 pKa = 11.84KK210 pKa = 10.27SSEE213 pKa = 3.71VVNRR217 pKa = 11.84LGEE220 pKa = 3.91IHH222 pKa = 6.71SLGRR226 pKa = 11.84HH227 pKa = 4.9GSSKK231 pKa = 10.07FVHH234 pKa = 6.09EE235 pKa = 4.69EE236 pKa = 3.06IRR238 pKa = 11.84YY239 pKa = 9.21KK240 pKa = 10.57RR241 pKa = 11.84GRR243 pKa = 11.84QKK245 pKa = 11.28
Molecular weight: 28.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11072412 |
25 |
5429 |
460.6 |
51.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.817 ± 0.013 | 1.861 ± 0.007 |
5.336 ± 0.01 | 6.552 ± 0.017 |
4.262 ± 0.01 | 6.463 ± 0.013 |
2.398 ± 0.006 | 5.39 ± 0.009 |
6.034 ± 0.015 | 9.739 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.36 ± 0.006 | 4.438 ± 0.009 |
4.835 ± 0.013 | 3.664 ± 0.01 |
5.462 ± 0.011 | 9.186 ± 0.019 |
4.716 ± 0.009 | 6.448 ± 0.011 |
1.264 ± 0.005 | 2.743 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
