
Grimontia hollisae CIP 101886
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Grimontia; Grimontia hollisae
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
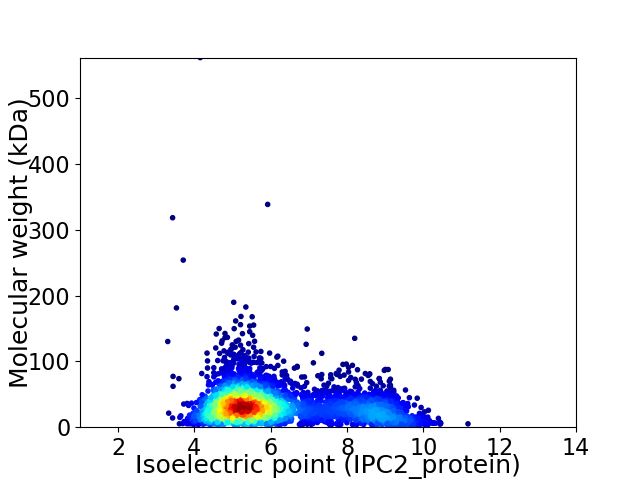
Virtual 2D-PAGE plot for 3553 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0IBX1|D0IBX1_GRIHO Putative 2-succinyl-6-hydroxy-2 4-cyclohexadiene-1-carboxylate synthase OS=Grimontia hollisae CIP 101886 OX=675812 GN=menH PE=3 SV=1
MM1 pKa = 7.62AVPALLAAGAAQASVNLYY19 pKa = 10.33DD20 pKa = 4.36ADD22 pKa = 3.91GVTVDD27 pKa = 4.23LSGAAEE33 pKa = 3.66IQYY36 pKa = 10.5FKK38 pKa = 10.81DD39 pKa = 3.49YY40 pKa = 10.59STTEE44 pKa = 3.84DD45 pKa = 3.31AYY47 pKa = 11.58LRR49 pKa = 11.84IDD51 pKa = 4.86DD52 pKa = 5.25ADD54 pKa = 4.18LLLTTSIEE62 pKa = 4.31VADD65 pKa = 4.5GLNAVAGMGFKK76 pKa = 11.0YY77 pKa = 10.27EE78 pKa = 4.2DD79 pKa = 3.36QFATGVTVSNDD90 pKa = 2.76NGTVKK95 pKa = 10.57NDD97 pKa = 3.26EE98 pKa = 4.74LYY100 pKa = 11.17VGLGGNFGTVTFGRR114 pKa = 11.84QLLLSDD120 pKa = 4.6DD121 pKa = 4.39AGNQKK126 pKa = 10.56DD127 pKa = 3.88YY128 pKa = 11.43EE129 pKa = 4.25LGYY132 pKa = 10.01EE133 pKa = 4.2QIDD136 pKa = 3.64FVQTEE141 pKa = 4.65GGQVIKK147 pKa = 10.0WVYY150 pKa = 11.45DD151 pKa = 3.05NGTFYY156 pKa = 11.22AGANVDD162 pKa = 3.87LDD164 pKa = 4.29TNKK167 pKa = 10.36SDD169 pKa = 3.56VTDD172 pKa = 3.54GRR174 pKa = 11.84QIIGGRR180 pKa = 11.84IGARR184 pKa = 11.84YY185 pKa = 9.04EE186 pKa = 4.05GLDD189 pKa = 3.3GRR191 pKa = 11.84IYY193 pKa = 10.44FYY195 pKa = 11.07DD196 pKa = 4.43GEE198 pKa = 4.57DD199 pKa = 3.74VDD201 pKa = 5.74GEE203 pKa = 4.42DD204 pKa = 3.18IKK206 pKa = 11.53GFNAEE211 pKa = 4.67LDD213 pKa = 3.44WTINDD218 pKa = 3.66QFDD221 pKa = 3.54LALSYY226 pKa = 11.08GQLDD230 pKa = 4.24YY231 pKa = 11.35EE232 pKa = 4.37LHH234 pKa = 6.07TNNAVGDD241 pKa = 4.01EE242 pKa = 4.04VDD244 pKa = 3.56VFGISAGYY252 pKa = 8.33QASEE256 pKa = 4.22KK257 pKa = 10.56LVFAIGYY264 pKa = 9.29DD265 pKa = 3.73YY266 pKa = 11.28LDD268 pKa = 3.91SKK270 pKa = 11.58GKK272 pKa = 10.52GGNLDD277 pKa = 3.52GDD279 pKa = 3.96ADD281 pKa = 4.23SIYY284 pKa = 11.47ANATYY289 pKa = 10.61KK290 pKa = 10.57LHH292 pKa = 6.39SNAMVYY298 pKa = 10.88AEE300 pKa = 4.53VGVADD305 pKa = 4.12GKK307 pKa = 11.01VGNVDD312 pKa = 2.76IDD314 pKa = 3.57TDD316 pKa = 3.43TGYY319 pKa = 11.56VLGMEE324 pKa = 4.63VKK326 pKa = 10.58FF327 pKa = 4.23
MM1 pKa = 7.62AVPALLAAGAAQASVNLYY19 pKa = 10.33DD20 pKa = 4.36ADD22 pKa = 3.91GVTVDD27 pKa = 4.23LSGAAEE33 pKa = 3.66IQYY36 pKa = 10.5FKK38 pKa = 10.81DD39 pKa = 3.49YY40 pKa = 10.59STTEE44 pKa = 3.84DD45 pKa = 3.31AYY47 pKa = 11.58LRR49 pKa = 11.84IDD51 pKa = 4.86DD52 pKa = 5.25ADD54 pKa = 4.18LLLTTSIEE62 pKa = 4.31VADD65 pKa = 4.5GLNAVAGMGFKK76 pKa = 11.0YY77 pKa = 10.27EE78 pKa = 4.2DD79 pKa = 3.36QFATGVTVSNDD90 pKa = 2.76NGTVKK95 pKa = 10.57NDD97 pKa = 3.26EE98 pKa = 4.74LYY100 pKa = 11.17VGLGGNFGTVTFGRR114 pKa = 11.84QLLLSDD120 pKa = 4.6DD121 pKa = 4.39AGNQKK126 pKa = 10.56DD127 pKa = 3.88YY128 pKa = 11.43EE129 pKa = 4.25LGYY132 pKa = 10.01EE133 pKa = 4.2QIDD136 pKa = 3.64FVQTEE141 pKa = 4.65GGQVIKK147 pKa = 10.0WVYY150 pKa = 11.45DD151 pKa = 3.05NGTFYY156 pKa = 11.22AGANVDD162 pKa = 3.87LDD164 pKa = 4.29TNKK167 pKa = 10.36SDD169 pKa = 3.56VTDD172 pKa = 3.54GRR174 pKa = 11.84QIIGGRR180 pKa = 11.84IGARR184 pKa = 11.84YY185 pKa = 9.04EE186 pKa = 4.05GLDD189 pKa = 3.3GRR191 pKa = 11.84IYY193 pKa = 10.44FYY195 pKa = 11.07DD196 pKa = 4.43GEE198 pKa = 4.57DD199 pKa = 3.74VDD201 pKa = 5.74GEE203 pKa = 4.42DD204 pKa = 3.18IKK206 pKa = 11.53GFNAEE211 pKa = 4.67LDD213 pKa = 3.44WTINDD218 pKa = 3.66QFDD221 pKa = 3.54LALSYY226 pKa = 11.08GQLDD230 pKa = 4.24YY231 pKa = 11.35EE232 pKa = 4.37LHH234 pKa = 6.07TNNAVGDD241 pKa = 4.01EE242 pKa = 4.04VDD244 pKa = 3.56VFGISAGYY252 pKa = 8.33QASEE256 pKa = 4.22KK257 pKa = 10.56LVFAIGYY264 pKa = 9.29DD265 pKa = 3.73YY266 pKa = 11.28LDD268 pKa = 3.91SKK270 pKa = 11.58GKK272 pKa = 10.52GGNLDD277 pKa = 3.52GDD279 pKa = 3.96ADD281 pKa = 4.23SIYY284 pKa = 11.47ANATYY289 pKa = 10.61KK290 pKa = 10.57LHH292 pKa = 6.39SNAMVYY298 pKa = 10.88AEE300 pKa = 4.53VGVADD305 pKa = 4.12GKK307 pKa = 11.01VGNVDD312 pKa = 2.76IDD314 pKa = 3.57TDD316 pKa = 3.43TGYY319 pKa = 11.56VLGMEE324 pKa = 4.63VKK326 pKa = 10.58FF327 pKa = 4.23
Molecular weight: 35.15 kDa
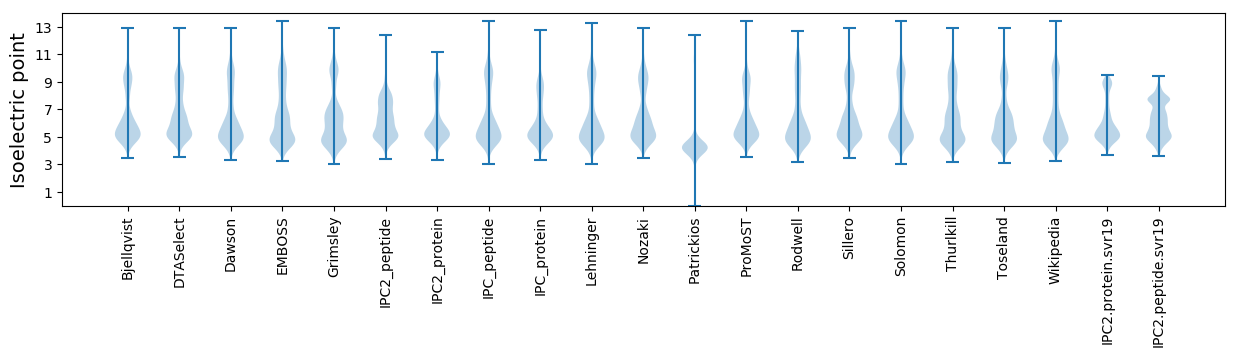
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0I2Z5|D0I2Z5_GRIHO Beta sliding clamp OS=Grimontia hollisae CIP 101886 OX=675812 GN=VHA_000108 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.44VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.33GRR39 pKa = 11.84SKK41 pKa = 10.8LSKK44 pKa = 10.52
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.44VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.33GRR39 pKa = 11.84SKK41 pKa = 10.8LSKK44 pKa = 10.52
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1131702 |
37 |
5261 |
318.5 |
35.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.267 ± 0.048 | 1.033 ± 0.015 |
5.688 ± 0.041 | 6.295 ± 0.041 |
4.006 ± 0.029 | 7.243 ± 0.043 |
2.232 ± 0.022 | 6.01 ± 0.034 |
4.981 ± 0.033 | 10.263 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.778 ± 0.023 | 4.018 ± 0.032 |
4.151 ± 0.028 | 4.066 ± 0.03 |
5.021 ± 0.042 | 6.102 ± 0.028 |
5.433 ± 0.046 | 7.338 ± 0.044 |
1.228 ± 0.016 | 2.849 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
