
Clostridium sp. FS41
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; unclassified Clostridium
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
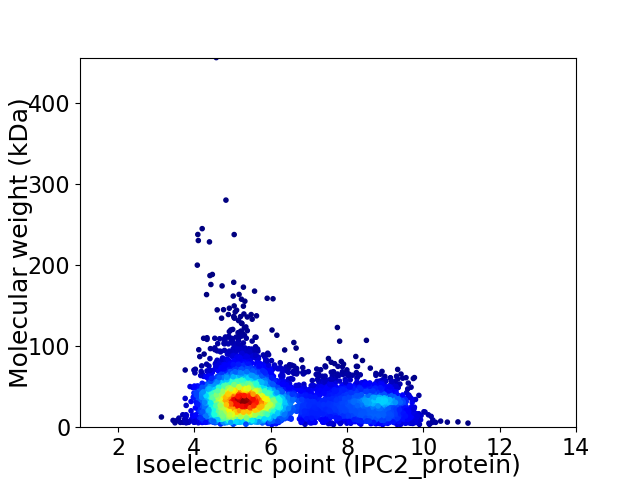
Virtual 2D-PAGE plot for 5722 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F0CK99|A0A0F0CK99_9CLOT Uncharacterized protein OS=Clostridium sp. FS41 OX=1609975 GN=CLFS41_12430 PE=4 SV=1
MM1 pKa = 7.25QMRR4 pKa = 11.84GCGSQGEE11 pKa = 4.46WVDD14 pKa = 3.61PYY16 pKa = 11.39KK17 pKa = 11.43VNTCFLDD24 pKa = 2.94TGYY27 pKa = 11.14FDD29 pKa = 3.34TCTFEE34 pKa = 3.83KK35 pKa = 10.29TPYY38 pKa = 10.41SNARR42 pKa = 11.84YY43 pKa = 8.73PYY45 pKa = 9.25QTVCDD50 pKa = 3.76NGLGLGYY57 pKa = 10.63YY58 pKa = 7.75EE59 pKa = 4.43VRR61 pKa = 11.84CPEE64 pKa = 3.81NSEE67 pKa = 4.27FDD69 pKa = 3.99PSTLRR74 pKa = 11.84CKK76 pKa = 10.33SVCEE80 pKa = 4.04YY81 pKa = 11.13GKK83 pKa = 10.26NPDD86 pKa = 3.94GTCMDD91 pKa = 3.88ACQFKK96 pKa = 10.92KK97 pKa = 10.76SIDD100 pKa = 4.02EE101 pKa = 4.15IKK103 pKa = 10.48SLQWLAYY110 pKa = 9.88VYY112 pKa = 10.64GEE114 pKa = 4.11QVTGSCYY121 pKa = 10.68GDD123 pKa = 3.56YY124 pKa = 11.09GATRR128 pKa = 11.84CEE130 pKa = 4.25LEE132 pKa = 4.18RR133 pKa = 11.84TPSDD137 pKa = 3.21STLCTGVDD145 pKa = 3.31SGQWTQNTICHH156 pKa = 6.32GNFQFTGNQCEE167 pKa = 4.28GGTLFWGKK175 pKa = 10.11DD176 pKa = 3.62GPDD179 pKa = 3.31TPIIPDD185 pKa = 4.37DD186 pKa = 5.14PIHH189 pKa = 7.61DD190 pKa = 4.88PDD192 pKa = 5.88DD193 pKa = 3.89PTGDD197 pKa = 3.64IEE199 pKa = 5.54DD200 pKa = 4.5PSILPDD206 pKa = 3.61GSTNTVNPPDD216 pKa = 4.02TDD218 pKa = 3.65SEE220 pKa = 4.63PDD222 pKa = 3.48VEE224 pKa = 5.55EE225 pKa = 5.22PDD227 pKa = 3.63TDD229 pKa = 3.49EE230 pKa = 4.45STDD233 pKa = 3.64TAVLKK238 pKa = 10.91AITGMNKK245 pKa = 10.0DD246 pKa = 3.38VNKK249 pKa = 10.54ALNDD253 pKa = 3.47MNIDD257 pKa = 3.4INQANADD264 pKa = 3.75VQNQIIALNASMVTNTQAIQKK285 pKa = 7.84QQINDD290 pKa = 3.14NKK292 pKa = 10.37IYY294 pKa = 10.49EE295 pKa = 4.16NTKK298 pKa = 10.95ALIQQANADD307 pKa = 3.49ITTAMNKK314 pKa = 7.52NTNAVNGVGDD324 pKa = 4.45DD325 pKa = 3.72VEE327 pKa = 5.55KK328 pKa = 10.74IAGAMDD334 pKa = 5.45GIAEE338 pKa = 4.26DD339 pKa = 3.65VSGISDD345 pKa = 3.9TLDD348 pKa = 4.1GIANTDD354 pKa = 3.08TSGAGTGGTCIEE366 pKa = 4.43SQSCTGFYY374 pKa = 10.69EE375 pKa = 4.04SGYY378 pKa = 9.68PDD380 pKa = 3.62GLGGLVSGQLDD391 pKa = 3.73DD392 pKa = 5.58LKK394 pKa = 11.35HH395 pKa = 4.84NTIDD399 pKa = 3.42NFVNSFGDD407 pKa = 3.73LDD409 pKa = 4.02LSSAKK414 pKa = 10.1RR415 pKa = 11.84PSFVLPVPFFGDD427 pKa = 3.76FSFEE431 pKa = 4.02EE432 pKa = 4.39QISFDD437 pKa = 3.32WVFGFIRR444 pKa = 11.84AVLIMTSVFAARR456 pKa = 11.84RR457 pKa = 11.84IIFGGG462 pKa = 3.39
MM1 pKa = 7.25QMRR4 pKa = 11.84GCGSQGEE11 pKa = 4.46WVDD14 pKa = 3.61PYY16 pKa = 11.39KK17 pKa = 11.43VNTCFLDD24 pKa = 2.94TGYY27 pKa = 11.14FDD29 pKa = 3.34TCTFEE34 pKa = 3.83KK35 pKa = 10.29TPYY38 pKa = 10.41SNARR42 pKa = 11.84YY43 pKa = 8.73PYY45 pKa = 9.25QTVCDD50 pKa = 3.76NGLGLGYY57 pKa = 10.63YY58 pKa = 7.75EE59 pKa = 4.43VRR61 pKa = 11.84CPEE64 pKa = 3.81NSEE67 pKa = 4.27FDD69 pKa = 3.99PSTLRR74 pKa = 11.84CKK76 pKa = 10.33SVCEE80 pKa = 4.04YY81 pKa = 11.13GKK83 pKa = 10.26NPDD86 pKa = 3.94GTCMDD91 pKa = 3.88ACQFKK96 pKa = 10.92KK97 pKa = 10.76SIDD100 pKa = 4.02EE101 pKa = 4.15IKK103 pKa = 10.48SLQWLAYY110 pKa = 9.88VYY112 pKa = 10.64GEE114 pKa = 4.11QVTGSCYY121 pKa = 10.68GDD123 pKa = 3.56YY124 pKa = 11.09GATRR128 pKa = 11.84CEE130 pKa = 4.25LEE132 pKa = 4.18RR133 pKa = 11.84TPSDD137 pKa = 3.21STLCTGVDD145 pKa = 3.31SGQWTQNTICHH156 pKa = 6.32GNFQFTGNQCEE167 pKa = 4.28GGTLFWGKK175 pKa = 10.11DD176 pKa = 3.62GPDD179 pKa = 3.31TPIIPDD185 pKa = 4.37DD186 pKa = 5.14PIHH189 pKa = 7.61DD190 pKa = 4.88PDD192 pKa = 5.88DD193 pKa = 3.89PTGDD197 pKa = 3.64IEE199 pKa = 5.54DD200 pKa = 4.5PSILPDD206 pKa = 3.61GSTNTVNPPDD216 pKa = 4.02TDD218 pKa = 3.65SEE220 pKa = 4.63PDD222 pKa = 3.48VEE224 pKa = 5.55EE225 pKa = 5.22PDD227 pKa = 3.63TDD229 pKa = 3.49EE230 pKa = 4.45STDD233 pKa = 3.64TAVLKK238 pKa = 10.91AITGMNKK245 pKa = 10.0DD246 pKa = 3.38VNKK249 pKa = 10.54ALNDD253 pKa = 3.47MNIDD257 pKa = 3.4INQANADD264 pKa = 3.75VQNQIIALNASMVTNTQAIQKK285 pKa = 7.84QQINDD290 pKa = 3.14NKK292 pKa = 10.37IYY294 pKa = 10.49EE295 pKa = 4.16NTKK298 pKa = 10.95ALIQQANADD307 pKa = 3.49ITTAMNKK314 pKa = 7.52NTNAVNGVGDD324 pKa = 4.45DD325 pKa = 3.72VEE327 pKa = 5.55KK328 pKa = 10.74IAGAMDD334 pKa = 5.45GIAEE338 pKa = 4.26DD339 pKa = 3.65VSGISDD345 pKa = 3.9TLDD348 pKa = 4.1GIANTDD354 pKa = 3.08TSGAGTGGTCIEE366 pKa = 4.43SQSCTGFYY374 pKa = 10.69EE375 pKa = 4.04SGYY378 pKa = 9.68PDD380 pKa = 3.62GLGGLVSGQLDD391 pKa = 3.73DD392 pKa = 5.58LKK394 pKa = 11.35HH395 pKa = 4.84NTIDD399 pKa = 3.42NFVNSFGDD407 pKa = 3.73LDD409 pKa = 4.02LSSAKK414 pKa = 10.1RR415 pKa = 11.84PSFVLPVPFFGDD427 pKa = 3.76FSFEE431 pKa = 4.02EE432 pKa = 4.39QISFDD437 pKa = 3.32WVFGFIRR444 pKa = 11.84AVLIMTSVFAARR456 pKa = 11.84RR457 pKa = 11.84IIFGGG462 pKa = 3.39
Molecular weight: 50.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F0CI08|A0A0F0CI08_9CLOT Orotate phosphoribosyltransferase OS=Clostridium sp. FS41 OX=1609975 GN=pyrE PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.44VHH16 pKa = 5.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.82TVGGRR28 pKa = 11.84KK29 pKa = 9.22VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.26GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.44VHH16 pKa = 5.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.82TVGGRR28 pKa = 11.84KK29 pKa = 9.22VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.26GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1853055 |
29 |
4083 |
323.8 |
36.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.891 ± 0.033 | 1.585 ± 0.013 |
5.587 ± 0.028 | 6.972 ± 0.035 |
4.023 ± 0.022 | 7.957 ± 0.031 |
1.792 ± 0.013 | 6.934 ± 0.029 |
5.646 ± 0.027 | 9.095 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.473 ± 0.016 | 3.868 ± 0.018 |
3.64 ± 0.022 | 3.396 ± 0.019 |
4.953 ± 0.029 | 5.908 ± 0.023 |
5.271 ± 0.023 | 6.99 ± 0.027 |
1.026 ± 0.012 | 3.991 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
