
Streptococcus varani
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
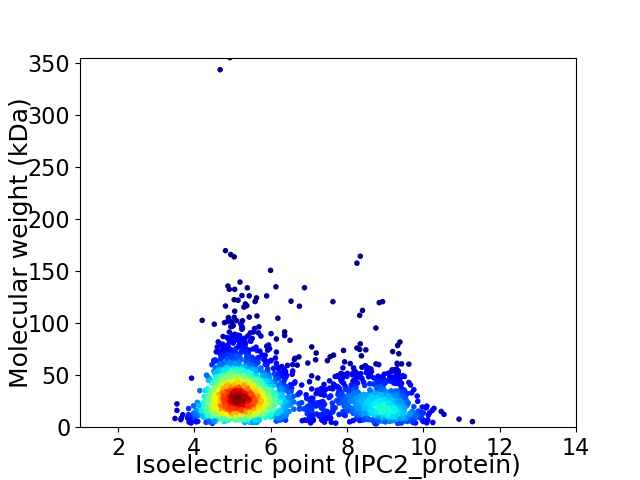
Virtual 2D-PAGE plot for 2422 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E4CTC7|A0A0E4CTC7_9STRE Sensor histidine kinase OS=Streptococcus varani OX=1608583 GN=BN1356_01945 PE=4 SV=1
MM1 pKa = 7.21NKK3 pKa = 9.55SEE5 pKa = 4.06KK6 pKa = 9.57MLYY9 pKa = 9.29TLEE12 pKa = 4.14QQDD15 pKa = 4.06LEE17 pKa = 4.15AAQAYY22 pKa = 9.79FEE24 pKa = 4.35EE25 pKa = 5.39ALLEE29 pKa = 4.61DD30 pKa = 5.12DD31 pKa = 5.81EE32 pKa = 4.57EE33 pKa = 4.35TLYY36 pKa = 11.27DD37 pKa = 3.8LGTYY41 pKa = 10.14LEE43 pKa = 4.96SIGFYY48 pKa = 10.02PQAKK52 pKa = 9.58EE53 pKa = 3.77IYY55 pKa = 8.47LQLVEE60 pKa = 5.55DD61 pKa = 4.71FPEE64 pKa = 4.94LYY66 pKa = 10.97LNLAQIANEE75 pKa = 4.15DD76 pKa = 3.79GAIEE80 pKa = 3.84EE81 pKa = 4.17AFAYY85 pKa = 10.43LEE87 pKa = 4.76NISKK91 pKa = 10.56SSPYY95 pKa = 9.9YY96 pKa = 10.75VEE98 pKa = 4.59ALLVKK103 pKa = 10.46ADD105 pKa = 4.7LYY107 pKa = 10.93QSEE110 pKa = 4.27GLADD114 pKa = 3.59VARR117 pKa = 11.84EE118 pKa = 3.93KK119 pKa = 11.03LLEE122 pKa = 4.31ATSLAEE128 pKa = 5.06DD129 pKa = 5.08PILTFGLAEE138 pKa = 4.8LDD140 pKa = 3.64MEE142 pKa = 4.9LGNFKK147 pKa = 10.31EE148 pKa = 4.63AIQGYY153 pKa = 7.93VQLDD157 pKa = 3.22NRR159 pKa = 11.84EE160 pKa = 4.08IYY162 pKa = 11.33DD163 pKa = 3.53MVGVSTYY170 pKa = 9.44QRR172 pKa = 11.84IGLAYY177 pKa = 9.37ASLGKK182 pKa = 10.44FEE184 pKa = 4.18VAIEE188 pKa = 4.0FLEE191 pKa = 4.26KK192 pKa = 10.42AVEE195 pKa = 4.31LEE197 pKa = 4.12YY198 pKa = 11.1DD199 pKa = 3.55DD200 pKa = 3.59QTVYY204 pKa = 11.03EE205 pKa = 4.77LATLLYY211 pKa = 10.55DD212 pKa = 4.58QEE214 pKa = 4.45EE215 pKa = 4.44CQKK218 pKa = 11.38ANLYY222 pKa = 9.66FKK224 pKa = 10.63QLDD227 pKa = 4.07TINPDD232 pKa = 3.27FEE234 pKa = 5.47GYY236 pKa = 10.07EE237 pKa = 3.83YY238 pKa = 10.81AYY240 pKa = 10.38AQSLHH245 pKa = 6.83AEE247 pKa = 4.28HH248 pKa = 7.12KK249 pKa = 10.46LDD251 pKa = 3.48EE252 pKa = 4.52ALAMVEE258 pKa = 4.29KK259 pKa = 10.78GLVKK263 pKa = 10.89NEE265 pKa = 3.79FDD267 pKa = 4.74SVLLLLASQYY277 pKa = 11.4AYY279 pKa = 10.3EE280 pKa = 4.12NHH282 pKa = 7.15DD283 pKa = 3.5NQAAEE288 pKa = 4.79DD289 pKa = 4.01YY290 pKa = 10.67LLKK293 pKa = 10.81AKK295 pKa = 10.31EE296 pKa = 4.57DD297 pKa = 3.95ADD299 pKa = 4.58DD300 pKa = 3.62MDD302 pKa = 5.23EE303 pKa = 4.99IILRR307 pKa = 11.84LTNLYY312 pKa = 10.46LEE314 pKa = 4.39QEE316 pKa = 4.16RR317 pKa = 11.84FDD319 pKa = 4.16AVVALASDD327 pKa = 4.08DD328 pKa = 4.13LDD330 pKa = 3.58NVLARR335 pKa = 11.84WNIAKK340 pKa = 10.09AYY342 pKa = 8.77QALEE346 pKa = 4.13EE347 pKa = 4.52DD348 pKa = 4.48EE349 pKa = 4.74KK350 pKa = 11.73ALDD353 pKa = 4.42LFDD356 pKa = 3.95EE357 pKa = 5.03LKK359 pKa = 10.98VEE361 pKa = 4.4LADD364 pKa = 3.94NPEE367 pKa = 3.78FLADD371 pKa = 3.52YY372 pKa = 10.27VQALKK377 pKa = 10.81QEE379 pKa = 4.46GSISEE384 pKa = 3.97ARR386 pKa = 11.84EE387 pKa = 3.43WAHH390 pKa = 7.11RR391 pKa = 11.84YY392 pKa = 8.53LHH394 pKa = 6.6LVPDD398 pKa = 5.53DD399 pKa = 3.69IAMQEE404 pKa = 4.39FYY406 pKa = 10.99NQEE409 pKa = 3.93DD410 pKa = 3.44
MM1 pKa = 7.21NKK3 pKa = 9.55SEE5 pKa = 4.06KK6 pKa = 9.57MLYY9 pKa = 9.29TLEE12 pKa = 4.14QQDD15 pKa = 4.06LEE17 pKa = 4.15AAQAYY22 pKa = 9.79FEE24 pKa = 4.35EE25 pKa = 5.39ALLEE29 pKa = 4.61DD30 pKa = 5.12DD31 pKa = 5.81EE32 pKa = 4.57EE33 pKa = 4.35TLYY36 pKa = 11.27DD37 pKa = 3.8LGTYY41 pKa = 10.14LEE43 pKa = 4.96SIGFYY48 pKa = 10.02PQAKK52 pKa = 9.58EE53 pKa = 3.77IYY55 pKa = 8.47LQLVEE60 pKa = 5.55DD61 pKa = 4.71FPEE64 pKa = 4.94LYY66 pKa = 10.97LNLAQIANEE75 pKa = 4.15DD76 pKa = 3.79GAIEE80 pKa = 3.84EE81 pKa = 4.17AFAYY85 pKa = 10.43LEE87 pKa = 4.76NISKK91 pKa = 10.56SSPYY95 pKa = 9.9YY96 pKa = 10.75VEE98 pKa = 4.59ALLVKK103 pKa = 10.46ADD105 pKa = 4.7LYY107 pKa = 10.93QSEE110 pKa = 4.27GLADD114 pKa = 3.59VARR117 pKa = 11.84EE118 pKa = 3.93KK119 pKa = 11.03LLEE122 pKa = 4.31ATSLAEE128 pKa = 5.06DD129 pKa = 5.08PILTFGLAEE138 pKa = 4.8LDD140 pKa = 3.64MEE142 pKa = 4.9LGNFKK147 pKa = 10.31EE148 pKa = 4.63AIQGYY153 pKa = 7.93VQLDD157 pKa = 3.22NRR159 pKa = 11.84EE160 pKa = 4.08IYY162 pKa = 11.33DD163 pKa = 3.53MVGVSTYY170 pKa = 9.44QRR172 pKa = 11.84IGLAYY177 pKa = 9.37ASLGKK182 pKa = 10.44FEE184 pKa = 4.18VAIEE188 pKa = 4.0FLEE191 pKa = 4.26KK192 pKa = 10.42AVEE195 pKa = 4.31LEE197 pKa = 4.12YY198 pKa = 11.1DD199 pKa = 3.55DD200 pKa = 3.59QTVYY204 pKa = 11.03EE205 pKa = 4.77LATLLYY211 pKa = 10.55DD212 pKa = 4.58QEE214 pKa = 4.45EE215 pKa = 4.44CQKK218 pKa = 11.38ANLYY222 pKa = 9.66FKK224 pKa = 10.63QLDD227 pKa = 4.07TINPDD232 pKa = 3.27FEE234 pKa = 5.47GYY236 pKa = 10.07EE237 pKa = 3.83YY238 pKa = 10.81AYY240 pKa = 10.38AQSLHH245 pKa = 6.83AEE247 pKa = 4.28HH248 pKa = 7.12KK249 pKa = 10.46LDD251 pKa = 3.48EE252 pKa = 4.52ALAMVEE258 pKa = 4.29KK259 pKa = 10.78GLVKK263 pKa = 10.89NEE265 pKa = 3.79FDD267 pKa = 4.74SVLLLLASQYY277 pKa = 11.4AYY279 pKa = 10.3EE280 pKa = 4.12NHH282 pKa = 7.15DD283 pKa = 3.5NQAAEE288 pKa = 4.79DD289 pKa = 4.01YY290 pKa = 10.67LLKK293 pKa = 10.81AKK295 pKa = 10.31EE296 pKa = 4.57DD297 pKa = 3.95ADD299 pKa = 4.58DD300 pKa = 3.62MDD302 pKa = 5.23EE303 pKa = 4.99IILRR307 pKa = 11.84LTNLYY312 pKa = 10.46LEE314 pKa = 4.39QEE316 pKa = 4.16RR317 pKa = 11.84FDD319 pKa = 4.16AVVALASDD327 pKa = 4.08DD328 pKa = 4.13LDD330 pKa = 3.58NVLARR335 pKa = 11.84WNIAKK340 pKa = 10.09AYY342 pKa = 8.77QALEE346 pKa = 4.13EE347 pKa = 4.52DD348 pKa = 4.48EE349 pKa = 4.74KK350 pKa = 11.73ALDD353 pKa = 4.42LFDD356 pKa = 3.95EE357 pKa = 5.03LKK359 pKa = 10.98VEE361 pKa = 4.4LADD364 pKa = 3.94NPEE367 pKa = 3.78FLADD371 pKa = 3.52YY372 pKa = 10.27VQALKK377 pKa = 10.81QEE379 pKa = 4.46GSISEE384 pKa = 3.97ARR386 pKa = 11.84EE387 pKa = 3.43WAHH390 pKa = 7.11RR391 pKa = 11.84YY392 pKa = 8.53LHH394 pKa = 6.6LVPDD398 pKa = 5.53DD399 pKa = 3.69IAMQEE404 pKa = 4.39FYY406 pKa = 10.99NQEE409 pKa = 3.93DD410 pKa = 3.44
Molecular weight: 47.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3WEJ7|A0A0E3WEJ7_9STRE Phage replication protein OS=Streptococcus varani OX=1608583 GN=BN1356_00167 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.26IRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.35HH16 pKa = 4.77GFRR19 pKa = 11.84NRR21 pKa = 11.84MATKK25 pKa = 9.7NGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.68GRR39 pKa = 11.84KK40 pKa = 8.47VLAVAA45 pKa = 4.94
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.26IRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.35HH16 pKa = 4.77GFRR19 pKa = 11.84NRR21 pKa = 11.84MATKK25 pKa = 9.7NGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.68GRR39 pKa = 11.84KK40 pKa = 8.47VLAVAA45 pKa = 4.94
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
711518 |
30 |
3228 |
293.8 |
32.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.585 ± 0.064 | 0.578 ± 0.015 |
5.713 ± 0.044 | 6.865 ± 0.047 |
4.602 ± 0.042 | 6.641 ± 0.05 |
1.977 ± 0.023 | 7.326 ± 0.053 |
6.683 ± 0.04 | 10.062 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.617 ± 0.025 | 4.407 ± 0.04 |
3.293 ± 0.025 | 4.187 ± 0.039 |
4.057 ± 0.035 | 6.324 ± 0.048 |
5.571 ± 0.055 | 6.754 ± 0.039 |
0.947 ± 0.017 | 3.811 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
