
Alces alces faeces associated microvirus MP15 5067
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.45
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2Z5CI08|A0A2Z5CI08_9VIRU Replication initiation protein OS=Alces alces faeces associated microvirus MP15 5067 OX=2219136 PE=4 SV=1
MM1 pKa = 7.27AVEE4 pKa = 4.38SKK6 pKa = 10.47IMRR9 pKa = 11.84GCCDD13 pKa = 3.17VFDD16 pKa = 4.58VSAHH20 pKa = 5.35YY21 pKa = 8.98PYY23 pKa = 9.93EE24 pKa = 4.22QNVGDD29 pKa = 4.11YY30 pKa = 10.51LVEE33 pKa = 4.65LGTYY37 pKa = 10.29LPLRR41 pKa = 11.84KK42 pKa = 9.33QLEE45 pKa = 4.11DD46 pKa = 3.48LKK48 pKa = 11.12RR49 pKa = 11.84AGVDD53 pKa = 3.29LRR55 pKa = 11.84AQRR58 pKa = 11.84EE59 pKa = 4.36ANSLYY64 pKa = 10.34QAGEE68 pKa = 4.35TIDD71 pKa = 3.75LTQEE75 pKa = 4.12YY76 pKa = 10.52VDD78 pKa = 4.17PLQVDD83 pKa = 4.11EE84 pKa = 5.54FDD86 pKa = 4.83LDD88 pKa = 3.93AKK90 pKa = 10.87RR91 pKa = 11.84EE92 pKa = 3.98LYY94 pKa = 10.67YY95 pKa = 11.17RR96 pKa = 11.84MIVEE100 pKa = 3.94QEE102 pKa = 3.81QARR105 pKa = 11.84KK106 pKa = 9.37SEE108 pKa = 4.23RR109 pKa = 11.84KK110 pKa = 9.29RR111 pKa = 11.84RR112 pKa = 11.84EE113 pKa = 3.58AEE115 pKa = 3.5FAEE118 pKa = 4.51FQKK121 pKa = 10.96QKK123 pKa = 10.47AQEE126 pKa = 4.16SAKK129 pKa = 10.53KK130 pKa = 10.57SVDD133 pKa = 3.21TTEE136 pKa = 4.26VDD138 pKa = 3.63SQQ140 pKa = 3.55
MM1 pKa = 7.27AVEE4 pKa = 4.38SKK6 pKa = 10.47IMRR9 pKa = 11.84GCCDD13 pKa = 3.17VFDD16 pKa = 4.58VSAHH20 pKa = 5.35YY21 pKa = 8.98PYY23 pKa = 9.93EE24 pKa = 4.22QNVGDD29 pKa = 4.11YY30 pKa = 10.51LVEE33 pKa = 4.65LGTYY37 pKa = 10.29LPLRR41 pKa = 11.84KK42 pKa = 9.33QLEE45 pKa = 4.11DD46 pKa = 3.48LKK48 pKa = 11.12RR49 pKa = 11.84AGVDD53 pKa = 3.29LRR55 pKa = 11.84AQRR58 pKa = 11.84EE59 pKa = 4.36ANSLYY64 pKa = 10.34QAGEE68 pKa = 4.35TIDD71 pKa = 3.75LTQEE75 pKa = 4.12YY76 pKa = 10.52VDD78 pKa = 4.17PLQVDD83 pKa = 4.11EE84 pKa = 5.54FDD86 pKa = 4.83LDD88 pKa = 3.93AKK90 pKa = 10.87RR91 pKa = 11.84EE92 pKa = 3.98LYY94 pKa = 10.67YY95 pKa = 11.17RR96 pKa = 11.84MIVEE100 pKa = 3.94QEE102 pKa = 3.81QARR105 pKa = 11.84KK106 pKa = 9.37SEE108 pKa = 4.23RR109 pKa = 11.84KK110 pKa = 9.29RR111 pKa = 11.84RR112 pKa = 11.84EE113 pKa = 3.58AEE115 pKa = 3.5FAEE118 pKa = 4.51FQKK121 pKa = 10.96QKK123 pKa = 10.47AQEE126 pKa = 4.16SAKK129 pKa = 10.53KK130 pKa = 10.57SVDD133 pKa = 3.21TTEE136 pKa = 4.26VDD138 pKa = 3.63SQQ140 pKa = 3.55
Molecular weight: 16.32 kDa
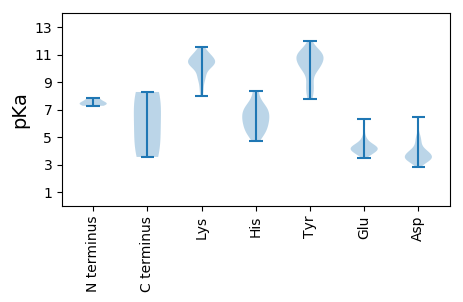
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJ78|A0A2Z5CJ78_9VIRU Major capsid protein OS=Alces alces faeces associated microvirus MP15 5067 OX=2219136 PE=3 SV=1
MM1 pKa = 7.49SCLSPTFIPVPANKK15 pKa = 10.27DD16 pKa = 3.4GVKK19 pKa = 10.41KK20 pKa = 10.58LVEE23 pKa = 3.87NPITHH28 pKa = 6.39KK29 pKa = 10.85LEE31 pKa = 3.99YY32 pKa = 10.91GMLVPCGKK40 pKa = 10.52CIPCKK45 pKa = 8.31VQKK48 pKa = 10.5ARR50 pKa = 11.84EE51 pKa = 3.65WSMRR55 pKa = 11.84CMLEE59 pKa = 3.77LPYY62 pKa = 10.08WYY64 pKa = 10.01DD65 pKa = 3.36ADD67 pKa = 4.8FVTLTYY73 pKa = 10.92DD74 pKa = 4.88DD75 pKa = 3.75EE76 pKa = 4.72HH77 pKa = 8.34LPEE80 pKa = 4.89NGSLVKK86 pKa = 10.26KK87 pKa = 10.63DD88 pKa = 3.16LTDD91 pKa = 3.81FFKK94 pKa = 10.79RR95 pKa = 11.84LRR97 pKa = 11.84RR98 pKa = 11.84DD99 pKa = 3.67LEE101 pKa = 4.11PLNIKK106 pKa = 10.16YY107 pKa = 7.73FACGEE112 pKa = 4.13YY113 pKa = 10.9GEE115 pKa = 4.45NTMRR119 pKa = 11.84PHH121 pKa = 4.7YY122 pKa = 9.75HH123 pKa = 6.96AIIFGVGLSEE133 pKa = 5.46RR134 pKa = 11.84EE135 pKa = 4.11NKK137 pKa = 10.35SLGCFYY143 pKa = 11.08SDD145 pKa = 4.33NNLIEE150 pKa = 4.96NNWPYY155 pKa = 11.75GNVFNGQVTPNSCRR169 pKa = 11.84YY170 pKa = 7.89CAEE173 pKa = 4.15YY174 pKa = 10.96VFKK177 pKa = 10.77KK178 pKa = 11.02YY179 pKa = 10.34NGKK182 pKa = 10.09KK183 pKa = 10.14KK184 pKa = 10.48IEE186 pKa = 4.18VYY188 pKa = 10.93DD189 pKa = 3.83NLGLQQPFQVQSNGIGKK206 pKa = 10.21RR207 pKa = 11.84YY208 pKa = 9.83LDD210 pKa = 3.98EE211 pKa = 4.36NMNDD215 pKa = 3.37LFQRR219 pKa = 11.84GYY221 pKa = 10.8VLYY224 pKa = 10.2HH225 pKa = 6.08GKK227 pKa = 9.65PYY229 pKa = 10.74SFPRR233 pKa = 11.84YY234 pKa = 8.91FADD237 pKa = 3.63KK238 pKa = 10.55LKK240 pKa = 11.37SNDD243 pKa = 3.19PQRR246 pKa = 11.84YY247 pKa = 8.82KK248 pKa = 11.2DD249 pKa = 3.69CLDD252 pKa = 3.99NIKK255 pKa = 10.62VGSYY259 pKa = 11.05EE260 pKa = 4.59DD261 pKa = 3.37FCEE264 pKa = 4.11DD265 pKa = 3.33VKK267 pKa = 10.99EE268 pKa = 4.42SEE270 pKa = 4.47MMSQYY275 pKa = 11.82DD276 pKa = 3.53LMQFYY281 pKa = 8.3KK282 pKa = 10.17TKK284 pKa = 10.35KK285 pKa = 9.31DD286 pKa = 3.29RR287 pKa = 11.84EE288 pKa = 4.22KK289 pKa = 10.87AIHH292 pKa = 6.9EE293 pKa = 4.42DD294 pKa = 3.88YY295 pKa = 10.78KK296 pKa = 11.53SKK298 pKa = 10.58FYY300 pKa = 11.29SNRR303 pKa = 11.84KK304 pKa = 8.28
MM1 pKa = 7.49SCLSPTFIPVPANKK15 pKa = 10.27DD16 pKa = 3.4GVKK19 pKa = 10.41KK20 pKa = 10.58LVEE23 pKa = 3.87NPITHH28 pKa = 6.39KK29 pKa = 10.85LEE31 pKa = 3.99YY32 pKa = 10.91GMLVPCGKK40 pKa = 10.52CIPCKK45 pKa = 8.31VQKK48 pKa = 10.5ARR50 pKa = 11.84EE51 pKa = 3.65WSMRR55 pKa = 11.84CMLEE59 pKa = 3.77LPYY62 pKa = 10.08WYY64 pKa = 10.01DD65 pKa = 3.36ADD67 pKa = 4.8FVTLTYY73 pKa = 10.92DD74 pKa = 4.88DD75 pKa = 3.75EE76 pKa = 4.72HH77 pKa = 8.34LPEE80 pKa = 4.89NGSLVKK86 pKa = 10.26KK87 pKa = 10.63DD88 pKa = 3.16LTDD91 pKa = 3.81FFKK94 pKa = 10.79RR95 pKa = 11.84LRR97 pKa = 11.84RR98 pKa = 11.84DD99 pKa = 3.67LEE101 pKa = 4.11PLNIKK106 pKa = 10.16YY107 pKa = 7.73FACGEE112 pKa = 4.13YY113 pKa = 10.9GEE115 pKa = 4.45NTMRR119 pKa = 11.84PHH121 pKa = 4.7YY122 pKa = 9.75HH123 pKa = 6.96AIIFGVGLSEE133 pKa = 5.46RR134 pKa = 11.84EE135 pKa = 4.11NKK137 pKa = 10.35SLGCFYY143 pKa = 11.08SDD145 pKa = 4.33NNLIEE150 pKa = 4.96NNWPYY155 pKa = 11.75GNVFNGQVTPNSCRR169 pKa = 11.84YY170 pKa = 7.89CAEE173 pKa = 4.15YY174 pKa = 10.96VFKK177 pKa = 10.77KK178 pKa = 11.02YY179 pKa = 10.34NGKK182 pKa = 10.09KK183 pKa = 10.14KK184 pKa = 10.48IEE186 pKa = 4.18VYY188 pKa = 10.93DD189 pKa = 3.83NLGLQQPFQVQSNGIGKK206 pKa = 10.21RR207 pKa = 11.84YY208 pKa = 9.83LDD210 pKa = 3.98EE211 pKa = 4.36NMNDD215 pKa = 3.37LFQRR219 pKa = 11.84GYY221 pKa = 10.8VLYY224 pKa = 10.2HH225 pKa = 6.08GKK227 pKa = 9.65PYY229 pKa = 10.74SFPRR233 pKa = 11.84YY234 pKa = 8.91FADD237 pKa = 3.63KK238 pKa = 10.55LKK240 pKa = 11.37SNDD243 pKa = 3.19PQRR246 pKa = 11.84YY247 pKa = 8.82KK248 pKa = 11.2DD249 pKa = 3.69CLDD252 pKa = 3.99NIKK255 pKa = 10.62VGSYY259 pKa = 11.05EE260 pKa = 4.59DD261 pKa = 3.37FCEE264 pKa = 4.11DD265 pKa = 3.33VKK267 pKa = 10.99EE268 pKa = 4.42SEE270 pKa = 4.47MMSQYY275 pKa = 11.82DD276 pKa = 3.53LMQFYY281 pKa = 8.3KK282 pKa = 10.17TKK284 pKa = 10.35KK285 pKa = 9.31DD286 pKa = 3.29RR287 pKa = 11.84EE288 pKa = 4.22KK289 pKa = 10.87AIHH292 pKa = 6.9EE293 pKa = 4.42DD294 pKa = 3.88YY295 pKa = 10.78KK296 pKa = 11.53SKK298 pKa = 10.58FYY300 pKa = 11.29SNRR303 pKa = 11.84KK304 pKa = 8.28
Molecular weight: 35.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1375 |
140 |
626 |
343.8 |
39.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.4 ± 2.486 | 1.382 ± 0.692 |
7.418 ± 0.636 | 6.764 ± 1.254 |
5.091 ± 1.174 | 5.527 ± 0.948 |
2.036 ± 0.241 | 3.709 ± 0.525 |
5.745 ± 1.627 | 7.636 ± 0.33 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.618 ± 0.259 | 5.6 ± 0.763 |
3.927 ± 0.794 | 4.727 ± 1.147 |
5.673 ± 0.439 | 7.127 ± 1.154 |
5.818 ± 1.176 | 6.255 ± 0.347 |
0.727 ± 0.22 | 5.818 ± 0.911 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
