
Sesamum indicum (Oriental sesame) (Sesamum orientale)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Pedaliaceae; Sesamum
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
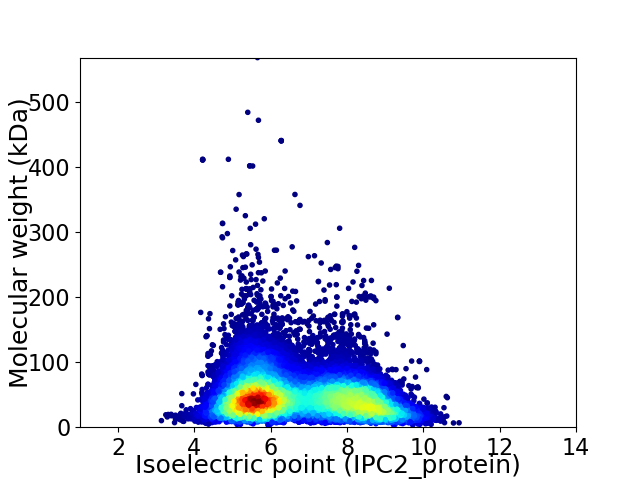
Virtual 2D-PAGE plot for 24106 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I9UEB5|A0A6I9UEB5_SESIN HEAT repeat-containing protein 5B OS=Sesamum indicum OX=4182 GN=LOC105177948 PE=4 SV=1
MM1 pKa = 7.16YY2 pKa = 10.63NSTRR6 pKa = 11.84LPNKK10 pKa = 9.45VFLISSFIFEE20 pKa = 4.64EE21 pKa = 4.49LEE23 pKa = 3.84KK24 pKa = 11.13DD25 pKa = 3.66AGGPCLGYY33 pKa = 10.62DD34 pKa = 3.67LQSLEE39 pKa = 4.06NLRR42 pKa = 11.84DD43 pKa = 3.79AEE45 pKa = 4.14EE46 pKa = 4.69LEE48 pKa = 4.15EE49 pKa = 6.23DD50 pKa = 4.48GGGPCLGYY58 pKa = 10.68DD59 pKa = 3.59PQRR62 pKa = 11.84LEE64 pKa = 3.89NLRR67 pKa = 11.84DD68 pKa = 3.75AEE70 pKa = 4.14EE71 pKa = 4.69LEE73 pKa = 4.15EE74 pKa = 6.23DD75 pKa = 4.48GGGPCLGYY83 pKa = 10.64DD84 pKa = 3.67LQSLEE89 pKa = 4.06NLRR92 pKa = 11.84DD93 pKa = 3.79AEE95 pKa = 4.14EE96 pKa = 4.69LEE98 pKa = 4.15EE99 pKa = 6.23DD100 pKa = 4.48GGGPCLGYY108 pKa = 10.67DD109 pKa = 3.64SQRR112 pKa = 11.84LEE114 pKa = 3.97NLRR117 pKa = 11.84DD118 pKa = 3.75AEE120 pKa = 4.25EE121 pKa = 4.78LEE123 pKa = 4.34EE124 pKa = 6.31DD125 pKa = 5.1DD126 pKa = 6.02GGPCLGYY133 pKa = 10.61DD134 pKa = 3.67LQSLEE139 pKa = 4.06NLRR142 pKa = 11.84DD143 pKa = 3.79AEE145 pKa = 4.14EE146 pKa = 4.69LEE148 pKa = 4.06EE149 pKa = 6.23DD150 pKa = 4.85GGGLCLGYY158 pKa = 10.61DD159 pKa = 3.68PQRR162 pKa = 11.84LEE164 pKa = 3.89NLRR167 pKa = 11.84DD168 pKa = 3.75AEE170 pKa = 4.14EE171 pKa = 4.69LEE173 pKa = 4.15EE174 pKa = 6.23DD175 pKa = 4.48GGGPCLGYY183 pKa = 10.68DD184 pKa = 3.59PQRR187 pKa = 11.84LEE189 pKa = 3.89NLRR192 pKa = 11.84DD193 pKa = 3.75AEE195 pKa = 4.25EE196 pKa = 4.78LEE198 pKa = 4.34EE199 pKa = 6.31DD200 pKa = 5.1DD201 pKa = 6.02GGPCLGYY208 pKa = 10.63DD209 pKa = 3.88LQRR212 pKa = 11.84LEE214 pKa = 4.17NLRR217 pKa = 11.84DD218 pKa = 3.75AEE220 pKa = 4.14EE221 pKa = 4.69LEE223 pKa = 4.15EE224 pKa = 6.23DD225 pKa = 4.48GGGPCLGYY233 pKa = 10.67DD234 pKa = 3.64SQRR237 pKa = 11.84LEE239 pKa = 3.97NLRR242 pKa = 11.84DD243 pKa = 3.75AEE245 pKa = 4.25EE246 pKa = 4.78LEE248 pKa = 4.34EE249 pKa = 6.31DD250 pKa = 5.1DD251 pKa = 6.02GGPCLGYY258 pKa = 10.63DD259 pKa = 3.88LQRR262 pKa = 11.84LEE264 pKa = 4.17NLRR267 pKa = 11.84DD268 pKa = 3.75AEE270 pKa = 4.25EE271 pKa = 4.78LEE273 pKa = 4.34EE274 pKa = 6.31DD275 pKa = 5.1DD276 pKa = 6.02GGPCLGYY283 pKa = 10.61DD284 pKa = 3.67LQSLEE289 pKa = 4.06NLRR292 pKa = 11.84DD293 pKa = 3.79AEE295 pKa = 4.14EE296 pKa = 4.69LEE298 pKa = 4.06EE299 pKa = 6.23DD300 pKa = 4.85GGGLCLGYY308 pKa = 10.61DD309 pKa = 3.68PQRR312 pKa = 11.84LEE314 pKa = 3.89NLRR317 pKa = 11.84DD318 pKa = 3.75AEE320 pKa = 4.25EE321 pKa = 4.78LEE323 pKa = 4.34EE324 pKa = 6.31DD325 pKa = 5.1DD326 pKa = 6.02GGPCLGYY333 pKa = 10.63DD334 pKa = 3.88LQRR337 pKa = 11.84LEE339 pKa = 4.17NLRR342 pKa = 11.84DD343 pKa = 3.75AEE345 pKa = 4.14EE346 pKa = 4.69LEE348 pKa = 4.15EE349 pKa = 6.23DD350 pKa = 4.48GGGPCLGYY358 pKa = 10.65DD359 pKa = 3.6PQSLEE364 pKa = 3.8NLRR367 pKa = 11.84DD368 pKa = 3.74AKK370 pKa = 10.66EE371 pKa = 3.99LEE373 pKa = 4.29EE374 pKa = 6.11DD375 pKa = 4.14DD376 pKa = 4.75GGPFLGYY383 pKa = 10.51DD384 pKa = 3.61LQSLEE389 pKa = 3.95NLRR392 pKa = 11.84DD393 pKa = 3.79EE394 pKa = 4.95EE395 pKa = 4.37EE396 pKa = 4.01LAEE399 pKa = 4.63DD400 pKa = 4.82DD401 pKa = 4.46RR402 pKa = 11.84GPSLGYY408 pKa = 10.8DD409 pKa = 3.7FLCPGKK415 pKa = 10.36LLAEE419 pKa = 4.41EE420 pKa = 4.56EE421 pKa = 4.49VQGDD425 pKa = 3.44SGGPCLGYY433 pKa = 10.2IGSTT437 pKa = 3.38
MM1 pKa = 7.16YY2 pKa = 10.63NSTRR6 pKa = 11.84LPNKK10 pKa = 9.45VFLISSFIFEE20 pKa = 4.64EE21 pKa = 4.49LEE23 pKa = 3.84KK24 pKa = 11.13DD25 pKa = 3.66AGGPCLGYY33 pKa = 10.62DD34 pKa = 3.67LQSLEE39 pKa = 4.06NLRR42 pKa = 11.84DD43 pKa = 3.79AEE45 pKa = 4.14EE46 pKa = 4.69LEE48 pKa = 4.15EE49 pKa = 6.23DD50 pKa = 4.48GGGPCLGYY58 pKa = 10.68DD59 pKa = 3.59PQRR62 pKa = 11.84LEE64 pKa = 3.89NLRR67 pKa = 11.84DD68 pKa = 3.75AEE70 pKa = 4.14EE71 pKa = 4.69LEE73 pKa = 4.15EE74 pKa = 6.23DD75 pKa = 4.48GGGPCLGYY83 pKa = 10.64DD84 pKa = 3.67LQSLEE89 pKa = 4.06NLRR92 pKa = 11.84DD93 pKa = 3.79AEE95 pKa = 4.14EE96 pKa = 4.69LEE98 pKa = 4.15EE99 pKa = 6.23DD100 pKa = 4.48GGGPCLGYY108 pKa = 10.67DD109 pKa = 3.64SQRR112 pKa = 11.84LEE114 pKa = 3.97NLRR117 pKa = 11.84DD118 pKa = 3.75AEE120 pKa = 4.25EE121 pKa = 4.78LEE123 pKa = 4.34EE124 pKa = 6.31DD125 pKa = 5.1DD126 pKa = 6.02GGPCLGYY133 pKa = 10.61DD134 pKa = 3.67LQSLEE139 pKa = 4.06NLRR142 pKa = 11.84DD143 pKa = 3.79AEE145 pKa = 4.14EE146 pKa = 4.69LEE148 pKa = 4.06EE149 pKa = 6.23DD150 pKa = 4.85GGGLCLGYY158 pKa = 10.61DD159 pKa = 3.68PQRR162 pKa = 11.84LEE164 pKa = 3.89NLRR167 pKa = 11.84DD168 pKa = 3.75AEE170 pKa = 4.14EE171 pKa = 4.69LEE173 pKa = 4.15EE174 pKa = 6.23DD175 pKa = 4.48GGGPCLGYY183 pKa = 10.68DD184 pKa = 3.59PQRR187 pKa = 11.84LEE189 pKa = 3.89NLRR192 pKa = 11.84DD193 pKa = 3.75AEE195 pKa = 4.25EE196 pKa = 4.78LEE198 pKa = 4.34EE199 pKa = 6.31DD200 pKa = 5.1DD201 pKa = 6.02GGPCLGYY208 pKa = 10.63DD209 pKa = 3.88LQRR212 pKa = 11.84LEE214 pKa = 4.17NLRR217 pKa = 11.84DD218 pKa = 3.75AEE220 pKa = 4.14EE221 pKa = 4.69LEE223 pKa = 4.15EE224 pKa = 6.23DD225 pKa = 4.48GGGPCLGYY233 pKa = 10.67DD234 pKa = 3.64SQRR237 pKa = 11.84LEE239 pKa = 3.97NLRR242 pKa = 11.84DD243 pKa = 3.75AEE245 pKa = 4.25EE246 pKa = 4.78LEE248 pKa = 4.34EE249 pKa = 6.31DD250 pKa = 5.1DD251 pKa = 6.02GGPCLGYY258 pKa = 10.63DD259 pKa = 3.88LQRR262 pKa = 11.84LEE264 pKa = 4.17NLRR267 pKa = 11.84DD268 pKa = 3.75AEE270 pKa = 4.25EE271 pKa = 4.78LEE273 pKa = 4.34EE274 pKa = 6.31DD275 pKa = 5.1DD276 pKa = 6.02GGPCLGYY283 pKa = 10.61DD284 pKa = 3.67LQSLEE289 pKa = 4.06NLRR292 pKa = 11.84DD293 pKa = 3.79AEE295 pKa = 4.14EE296 pKa = 4.69LEE298 pKa = 4.06EE299 pKa = 6.23DD300 pKa = 4.85GGGLCLGYY308 pKa = 10.61DD309 pKa = 3.68PQRR312 pKa = 11.84LEE314 pKa = 3.89NLRR317 pKa = 11.84DD318 pKa = 3.75AEE320 pKa = 4.25EE321 pKa = 4.78LEE323 pKa = 4.34EE324 pKa = 6.31DD325 pKa = 5.1DD326 pKa = 6.02GGPCLGYY333 pKa = 10.63DD334 pKa = 3.88LQRR337 pKa = 11.84LEE339 pKa = 4.17NLRR342 pKa = 11.84DD343 pKa = 3.75AEE345 pKa = 4.14EE346 pKa = 4.69LEE348 pKa = 4.15EE349 pKa = 6.23DD350 pKa = 4.48GGGPCLGYY358 pKa = 10.65DD359 pKa = 3.6PQSLEE364 pKa = 3.8NLRR367 pKa = 11.84DD368 pKa = 3.74AKK370 pKa = 10.66EE371 pKa = 3.99LEE373 pKa = 4.29EE374 pKa = 6.11DD375 pKa = 4.14DD376 pKa = 4.75GGPFLGYY383 pKa = 10.51DD384 pKa = 3.61LQSLEE389 pKa = 3.95NLRR392 pKa = 11.84DD393 pKa = 3.79EE394 pKa = 4.95EE395 pKa = 4.37EE396 pKa = 4.01LAEE399 pKa = 4.63DD400 pKa = 4.82DD401 pKa = 4.46RR402 pKa = 11.84GPSLGYY408 pKa = 10.8DD409 pKa = 3.7FLCPGKK415 pKa = 10.36LLAEE419 pKa = 4.41EE420 pKa = 4.56EE421 pKa = 4.49VQGDD425 pKa = 3.44SGGPCLGYY433 pKa = 10.2IGSTT437 pKa = 3.38
Molecular weight: 48.31 kDa
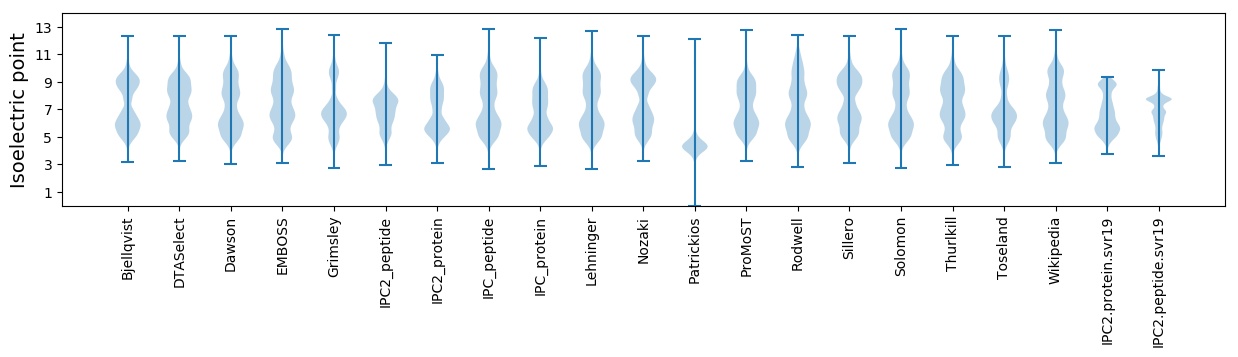
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I9UHL4|A0A6I9UHL4_SESIN MADS-box protein SOC1 isoform X2 OS=Sesamum indicum OX=4182 GN=LOC105179249 PE=4 SV=1
MM1 pKa = 7.39AAAGSEE7 pKa = 4.08LGAKK11 pKa = 9.27EE12 pKa = 4.0VGSVLRR18 pKa = 11.84SHH20 pKa = 6.97KK21 pKa = 10.44GAGRR25 pKa = 11.84GVAILDD31 pKa = 4.29FILRR35 pKa = 11.84LVGIVATLASAIAMATTNEE54 pKa = 4.17SLPFFTQFIRR64 pKa = 11.84FRR66 pKa = 11.84AKK68 pKa = 10.95YY69 pKa = 9.92NDD71 pKa = 3.74LPTFTFFVAANSVVCAYY88 pKa = 10.6LVLSLALSIFHH99 pKa = 7.09IVRR102 pKa = 11.84SSAQNSRR109 pKa = 11.84VVLIFFDD116 pKa = 3.81TGMLGLLSAGASASAAIVYY135 pKa = 8.38LAHH138 pKa = 7.37KK139 pKa = 10.58GNTRR143 pKa = 11.84ANWFAICQQFNSFCQRR159 pKa = 11.84TSGSLIGSFGGMIVLVLLILMSAANLSRR187 pKa = 11.84SRR189 pKa = 3.69
MM1 pKa = 7.39AAAGSEE7 pKa = 4.08LGAKK11 pKa = 9.27EE12 pKa = 4.0VGSVLRR18 pKa = 11.84SHH20 pKa = 6.97KK21 pKa = 10.44GAGRR25 pKa = 11.84GVAILDD31 pKa = 4.29FILRR35 pKa = 11.84LVGIVATLASAIAMATTNEE54 pKa = 4.17SLPFFTQFIRR64 pKa = 11.84FRR66 pKa = 11.84AKK68 pKa = 10.95YY69 pKa = 9.92NDD71 pKa = 3.74LPTFTFFVAANSVVCAYY88 pKa = 10.6LVLSLALSIFHH99 pKa = 7.09IVRR102 pKa = 11.84SSAQNSRR109 pKa = 11.84VVLIFFDD116 pKa = 3.81TGMLGLLSAGASASAAIVYY135 pKa = 8.38LAHH138 pKa = 7.37KK139 pKa = 10.58GNTRR143 pKa = 11.84ANWFAICQQFNSFCQRR159 pKa = 11.84TSGSLIGSFGGMIVLVLLILMSAANLSRR187 pKa = 11.84SRR189 pKa = 3.69
Molecular weight: 20.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10991492 |
29 |
5106 |
456.0 |
50.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.031 ± 0.013 | 1.812 ± 0.007 |
5.351 ± 0.011 | 6.545 ± 0.017 |
4.094 ± 0.009 | 6.587 ± 0.014 |
2.383 ± 0.007 | 5.228 ± 0.009 |
6.025 ± 0.015 | 9.62 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.411 ± 0.006 | 4.347 ± 0.008 |
5.032 ± 0.012 | 3.71 ± 0.01 |
5.474 ± 0.012 | 8.923 ± 0.02 |
4.807 ± 0.008 | 6.527 ± 0.009 |
1.272 ± 0.005 | 2.811 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
