
Flavobacterium cyanobacteriorum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
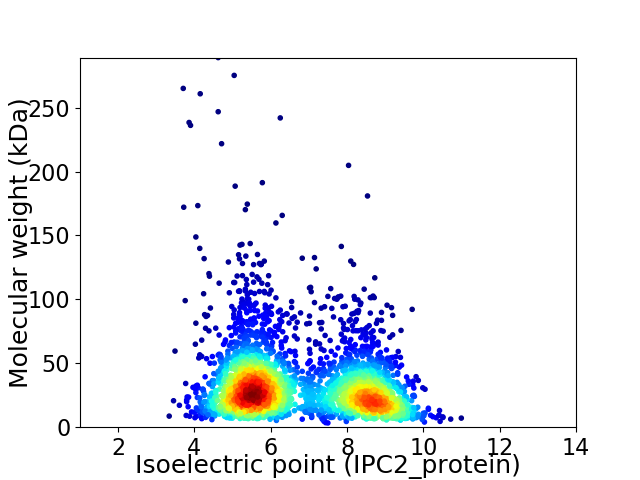
Virtual 2D-PAGE plot for 2995 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
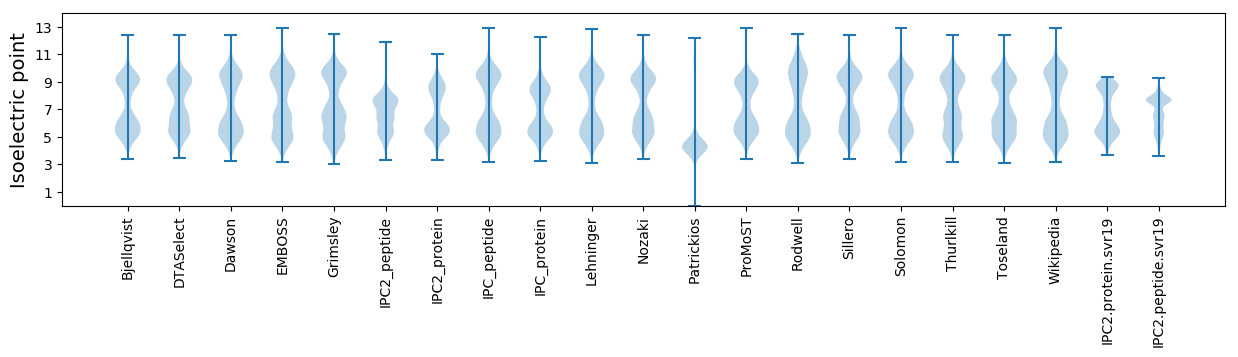
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A256A2H5|A0A256A2H5_9FLAO Uncharacterized protein OS=Flavobacterium cyanobacteriorum OX=2022802 GN=CHU92_00935 PE=4 SV=1
MM1 pKa = 7.5ALSVYY6 pKa = 10.23VISCSPDD13 pKa = 3.24NNNGEE18 pKa = 4.28APPRR22 pKa = 11.84DD23 pKa = 3.75YY24 pKa = 11.34AVQYY28 pKa = 10.86ASEE31 pKa = 4.28KK32 pKa = 10.92ADD34 pKa = 3.23IEE36 pKa = 4.27NFLKK40 pKa = 9.1KK41 pKa = 9.17TYY43 pKa = 9.68MVVDD47 pKa = 3.89EE48 pKa = 4.59TTWDD52 pKa = 3.63VVIDD56 pKa = 4.3SIGPDD61 pKa = 3.56TPQVSIWDD69 pKa = 3.55QTEE72 pKa = 3.84YY73 pKa = 10.72PLQQKK78 pKa = 9.25IVRR81 pKa = 11.84SNNVDD86 pKa = 3.22YY87 pKa = 10.03TVYY90 pKa = 10.92YY91 pKa = 8.76ITFNEE96 pKa = 4.92GVGSAPIKK104 pKa = 10.29ADD106 pKa = 3.12NVFISYY112 pKa = 10.37RR113 pKa = 11.84GIRR116 pKa = 11.84LDD118 pKa = 3.74TEE120 pKa = 3.87QFTYY124 pKa = 11.03VPFPANYY131 pKa = 9.9SSLLSTIEE139 pKa = 3.79GWQEE143 pKa = 3.66IIPLFKK149 pKa = 10.86AGIEE153 pKa = 4.51TNTNPQDD160 pKa = 3.51PASFSDD166 pKa = 3.7YY167 pKa = 10.81GAGVMFLPSGLGYY180 pKa = 10.92YY181 pKa = 10.23NISRR185 pKa = 11.84VGIPSYY191 pKa = 11.14SSLIFSFKK199 pKa = 10.61LFAIQRR205 pKa = 11.84ADD207 pKa = 3.39NDD209 pKa = 3.81RR210 pKa = 11.84DD211 pKa = 3.62GVLSVNEE218 pKa = 4.1TGGFADD224 pKa = 4.2IDD226 pKa = 4.71DD227 pKa = 4.7YY228 pKa = 12.15DD229 pKa = 5.78SDD231 pKa = 5.52DD232 pKa = 5.65DD233 pKa = 6.12DD234 pKa = 3.53IANYY238 pKa = 10.41RR239 pKa = 11.84DD240 pKa = 3.39VDD242 pKa = 4.04DD243 pKa = 6.03DD244 pKa = 4.35NDD246 pKa = 4.9GILTKK251 pKa = 10.42NEE253 pKa = 3.94KK254 pKa = 10.17QGTGSPEE261 pKa = 4.81DD262 pKa = 4.59LDD264 pKa = 4.01TDD266 pKa = 3.4NDD268 pKa = 4.81GIKK271 pKa = 10.58NYY273 pKa = 10.55LDD275 pKa = 3.71TDD277 pKa = 4.13DD278 pKa = 6.02DD279 pKa = 5.19GDD281 pKa = 4.6GIPTKK286 pKa = 10.8DD287 pKa = 3.91EE288 pKa = 4.54LNLPPCHH295 pKa = 6.68NNPAVPRR302 pKa = 11.84YY303 pKa = 10.02LNSSCC308 pKa = 5.02
MM1 pKa = 7.5ALSVYY6 pKa = 10.23VISCSPDD13 pKa = 3.24NNNGEE18 pKa = 4.28APPRR22 pKa = 11.84DD23 pKa = 3.75YY24 pKa = 11.34AVQYY28 pKa = 10.86ASEE31 pKa = 4.28KK32 pKa = 10.92ADD34 pKa = 3.23IEE36 pKa = 4.27NFLKK40 pKa = 9.1KK41 pKa = 9.17TYY43 pKa = 9.68MVVDD47 pKa = 3.89EE48 pKa = 4.59TTWDD52 pKa = 3.63VVIDD56 pKa = 4.3SIGPDD61 pKa = 3.56TPQVSIWDD69 pKa = 3.55QTEE72 pKa = 3.84YY73 pKa = 10.72PLQQKK78 pKa = 9.25IVRR81 pKa = 11.84SNNVDD86 pKa = 3.22YY87 pKa = 10.03TVYY90 pKa = 10.92YY91 pKa = 8.76ITFNEE96 pKa = 4.92GVGSAPIKK104 pKa = 10.29ADD106 pKa = 3.12NVFISYY112 pKa = 10.37RR113 pKa = 11.84GIRR116 pKa = 11.84LDD118 pKa = 3.74TEE120 pKa = 3.87QFTYY124 pKa = 11.03VPFPANYY131 pKa = 9.9SSLLSTIEE139 pKa = 3.79GWQEE143 pKa = 3.66IIPLFKK149 pKa = 10.86AGIEE153 pKa = 4.51TNTNPQDD160 pKa = 3.51PASFSDD166 pKa = 3.7YY167 pKa = 10.81GAGVMFLPSGLGYY180 pKa = 10.92YY181 pKa = 10.23NISRR185 pKa = 11.84VGIPSYY191 pKa = 11.14SSLIFSFKK199 pKa = 10.61LFAIQRR205 pKa = 11.84ADD207 pKa = 3.39NDD209 pKa = 3.81RR210 pKa = 11.84DD211 pKa = 3.62GVLSVNEE218 pKa = 4.1TGGFADD224 pKa = 4.2IDD226 pKa = 4.71DD227 pKa = 4.7YY228 pKa = 12.15DD229 pKa = 5.78SDD231 pKa = 5.52DD232 pKa = 5.65DD233 pKa = 6.12DD234 pKa = 3.53IANYY238 pKa = 10.41RR239 pKa = 11.84DD240 pKa = 3.39VDD242 pKa = 4.04DD243 pKa = 6.03DD244 pKa = 4.35NDD246 pKa = 4.9GILTKK251 pKa = 10.42NEE253 pKa = 3.94KK254 pKa = 10.17QGTGSPEE261 pKa = 4.81DD262 pKa = 4.59LDD264 pKa = 4.01TDD266 pKa = 3.4NDD268 pKa = 4.81GIKK271 pKa = 10.58NYY273 pKa = 10.55LDD275 pKa = 3.71TDD277 pKa = 4.13DD278 pKa = 6.02DD279 pKa = 5.19GDD281 pKa = 4.6GIPTKK286 pKa = 10.8DD287 pKa = 3.91EE288 pKa = 4.54LNLPPCHH295 pKa = 6.68NNPAVPRR302 pKa = 11.84YY303 pKa = 10.02LNSSCC308 pKa = 5.02
Molecular weight: 34.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A256A801|A0A256A801_9FLAO HTH araC/xylS-type domain-containing protein OS=Flavobacterium cyanobacteriorum OX=2022802 GN=CHU92_00355 PE=4 SV=1
AA1 pKa = 7.44LRR3 pKa = 11.84AFRR6 pKa = 11.84CNPSRR11 pKa = 11.84GVGGEE16 pKa = 3.73RR17 pKa = 11.84GVGSFHH23 pKa = 8.0KK24 pKa = 10.68YY25 pKa = 9.44LLKK28 pKa = 10.65ININPLYY35 pKa = 9.17IWFVVLEE42 pKa = 4.29IPSGFPHH49 pKa = 6.81NAPAGLPTASSSPPSGRR66 pKa = 11.84GCRR69 pKa = 11.84PHH71 pKa = 6.29HH72 pKa = 5.92TAFLSPIAPPP82 pKa = 3.62
AA1 pKa = 7.44LRR3 pKa = 11.84AFRR6 pKa = 11.84CNPSRR11 pKa = 11.84GVGGEE16 pKa = 3.73RR17 pKa = 11.84GVGSFHH23 pKa = 8.0KK24 pKa = 10.68YY25 pKa = 9.44LLKK28 pKa = 10.65ININPLYY35 pKa = 9.17IWFVVLEE42 pKa = 4.29IPSGFPHH49 pKa = 6.81NAPAGLPTASSSPPSGRR66 pKa = 11.84GCRR69 pKa = 11.84PHH71 pKa = 6.29HH72 pKa = 5.92TAFLSPIAPPP82 pKa = 3.62
Molecular weight: 8.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
945582 |
29 |
2789 |
315.7 |
35.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.258 ± 0.047 | 0.799 ± 0.016 |
5.244 ± 0.034 | 6.357 ± 0.067 |
5.128 ± 0.041 | 6.653 ± 0.052 |
1.779 ± 0.025 | 7.469 ± 0.042 |
7.21 ± 0.083 | 9.12 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.225 ± 0.029 | 5.873 ± 0.056 |
3.736 ± 0.032 | 3.468 ± 0.025 |
4.004 ± 0.033 | 6.024 ± 0.044 |
5.978 ± 0.088 | 6.425 ± 0.032 |
1.011 ± 0.019 | 4.235 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
