
Alteromonadales bacterium alter-6D02
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; unclassified Alteromonadales
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
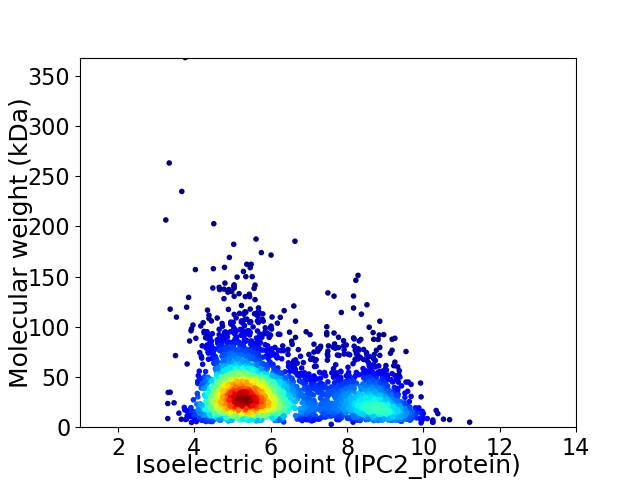
Virtual 2D-PAGE plot for 3415 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
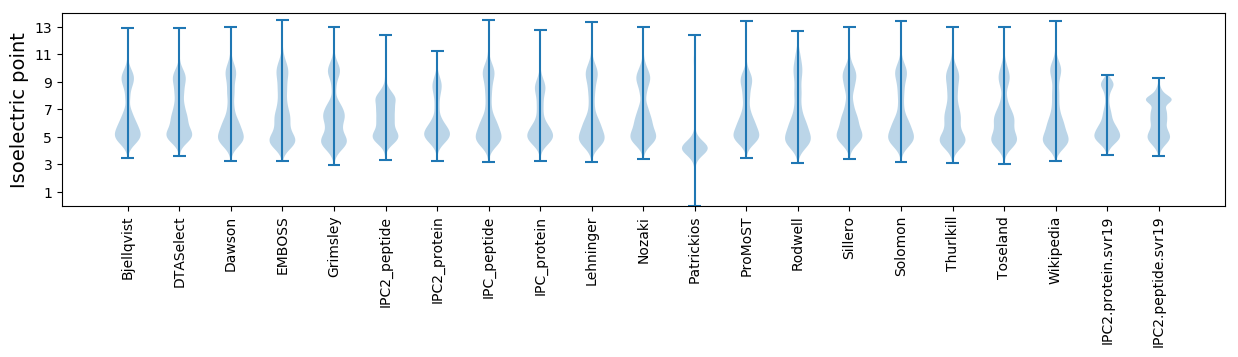
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N0WML2|A0A2N0WML2_9GAMM Chain length determinant family protein OS=Alteromonadales bacterium alter-6D02 OX=2058087 GN=CW748_14130 PE=4 SV=1
MM1 pKa = 7.05NVAMNTPPTPGAQTPAQEE19 pKa = 4.41VIKK22 pKa = 10.27VTGQTPLTNDD32 pKa = 2.97QQGPYY37 pKa = 10.3LLGVAQKK44 pKa = 8.56ITAEE48 pKa = 4.12EE49 pKa = 4.21IEE51 pKa = 4.31RR52 pKa = 11.84SNAKK56 pKa = 8.41TLTQQLKK63 pKa = 8.29NTLVSVNINDD73 pKa = 3.59VQNNPFQPDD82 pKa = 3.13VQYY85 pKa = 11.16RR86 pKa = 11.84GFTASPLLGLPQGISVYY103 pKa = 10.87LDD105 pKa = 3.38GIRR108 pKa = 11.84FNEE111 pKa = 4.1PFGDD115 pKa = 3.78TVNWDD120 pKa = 4.91LIALTAIDD128 pKa = 4.72NVTLYY133 pKa = 10.75SGSNPVFGQNTLGGALALSVKK154 pKa = 10.71DD155 pKa = 3.22GFTYY159 pKa = 10.51DD160 pKa = 3.31GHH162 pKa = 6.28EE163 pKa = 4.57LSAQAGSFGEE173 pKa = 4.18KK174 pKa = 9.63QLNVQSGGTSGNWGYY189 pKa = 11.18YY190 pKa = 10.49LNLNHH195 pKa = 6.39YY196 pKa = 9.86EE197 pKa = 3.87EE198 pKa = 5.67DD199 pKa = 3.21GWRR202 pKa = 11.84DD203 pKa = 3.43GSPSEE208 pKa = 4.23VKK210 pKa = 10.41QYY212 pKa = 10.95LANLSYY218 pKa = 9.75KK219 pKa = 10.06TDD221 pKa = 4.07DD222 pKa = 3.6LTADD226 pKa = 4.43LLFATNNNTMIGNGTVPIEE245 pKa = 4.05LLEE248 pKa = 4.25QEE250 pKa = 4.7SSTAIYY256 pKa = 7.54TQPDD260 pKa = 3.38QTKK263 pKa = 9.68TNLNMLGLNITAALTEE279 pKa = 4.47DD280 pKa = 3.2SSLAVNAYY288 pKa = 8.37YY289 pKa = 10.49RR290 pKa = 11.84KK291 pKa = 10.31NEE293 pKa = 3.65ISSINGDD300 pKa = 3.36DD301 pKa = 3.65SDD303 pKa = 4.4YY304 pKa = 11.64DD305 pKa = 3.53EE306 pKa = 6.24CEE308 pKa = 3.89FGTAVTLCEE317 pKa = 5.32GEE319 pKa = 5.4DD320 pKa = 4.35DD321 pKa = 6.08DD322 pKa = 7.0DD323 pKa = 6.76DD324 pKa = 5.51EE325 pKa = 6.44IDD327 pKa = 4.48GIEE330 pKa = 4.15SHH332 pKa = 7.04GDD334 pKa = 3.26DD335 pKa = 4.33DD336 pKa = 6.32DD337 pKa = 4.51EE338 pKa = 6.59GEE340 pKa = 4.39EE341 pKa = 3.99IEE343 pKa = 4.29PVHH346 pKa = 5.71FVGYY350 pKa = 10.81NEE352 pKa = 4.11GQYY355 pKa = 11.08LSDD358 pKa = 4.41IMPALDD364 pKa = 4.94ADD366 pKa = 5.45DD367 pKa = 5.22IDD369 pKa = 3.87GTYY372 pKa = 8.99NTGATDD378 pKa = 3.48NEE380 pKa = 4.68SFGFSAQLVTQWGNGHH396 pKa = 5.99EE397 pKa = 4.34LIIGGGADD405 pKa = 3.17KK406 pKa = 11.26ADD408 pKa = 3.08ISFASDD414 pKa = 3.1TQFAILHH421 pKa = 6.48NDD423 pKa = 3.01TAEE426 pKa = 4.72DD427 pKa = 3.89DD428 pKa = 4.3RR429 pKa = 11.84SVTPIPGLFDD439 pKa = 3.27QEE441 pKa = 4.46STVRR445 pKa = 11.84LDD447 pKa = 3.6TEE449 pKa = 4.02TTQYY453 pKa = 11.42YY454 pKa = 9.84FYY456 pKa = 10.7LANRR460 pKa = 11.84FKK462 pKa = 10.86LASDD466 pKa = 3.85LHH468 pKa = 8.15LDD470 pKa = 3.15IAGRR474 pKa = 11.84YY475 pKa = 8.0HH476 pKa = 8.01DD477 pKa = 5.91SNILMNDD484 pKa = 4.39LIDD487 pKa = 4.86DD488 pKa = 4.54GEE490 pKa = 4.9GSLDD494 pKa = 3.57GDD496 pKa = 3.69HH497 pKa = 7.4DD498 pKa = 4.63FSRR501 pKa = 11.84FNPAIGLSYY510 pKa = 10.45QISPQLTAKK519 pKa = 10.63LSYY522 pKa = 10.38SEE524 pKa = 4.61SSRR527 pKa = 11.84NPSPAEE533 pKa = 3.73LSCADD538 pKa = 4.11EE539 pKa = 5.82DD540 pKa = 5.48DD541 pKa = 4.21PCKK544 pKa = 10.66LPNGFVSDD552 pKa = 4.38PPLKK556 pKa = 10.01QVVVNTVEE564 pKa = 4.64LSLLGKK570 pKa = 10.04DD571 pKa = 4.18DD572 pKa = 3.77NLNYY576 pKa = 10.67GVTFFSTQSQDD587 pKa = 4.97DD588 pKa = 4.39IIFQQAGEE596 pKa = 4.29HH597 pKa = 6.43PARR600 pKa = 11.84GYY602 pKa = 10.03FVNIDD607 pKa = 3.17EE608 pKa = 4.5TQRR611 pKa = 11.84RR612 pKa = 11.84GVEE615 pKa = 3.87LSASYY620 pKa = 10.25RR621 pKa = 11.84GEE623 pKa = 4.07SFDD626 pKa = 3.21ITSSYY631 pKa = 11.36NYY633 pKa = 10.42LDD635 pKa = 3.62ATFEE639 pKa = 4.54SPFVSFSPVNPLGPNRR655 pKa = 11.84QVSPGDD661 pKa = 3.8TIPGQPQHH669 pKa = 5.9QIKK672 pKa = 10.95LIGQYY677 pKa = 9.76QATEE681 pKa = 4.03SFDD684 pKa = 3.26IGSEE688 pKa = 3.8LSYY691 pKa = 11.2ASEE694 pKa = 4.02SYY696 pKa = 10.98YY697 pKa = 10.92RR698 pKa = 11.84GDD700 pKa = 3.39EE701 pKa = 4.12ANEE704 pKa = 3.7NQQIGSHH711 pKa = 4.7TVVNLFANYY720 pKa = 10.17QITQQLALEE729 pKa = 4.26LRR731 pKa = 11.84VDD733 pKa = 3.77NVFDD737 pKa = 3.55RR738 pKa = 11.84QYY740 pKa = 10.37FTFGAYY746 pKa = 10.04GEE748 pKa = 4.17ADD750 pKa = 3.05EE751 pKa = 5.0VLEE754 pKa = 4.08NVYY757 pKa = 10.63PDD759 pKa = 3.35IEE761 pKa = 4.38DD762 pKa = 4.08PYY764 pKa = 11.18FVGPAKK770 pKa = 9.99PRR772 pKa = 11.84SVSVEE777 pKa = 3.63LSYY780 pKa = 11.19QFF782 pKa = 4.51
MM1 pKa = 7.05NVAMNTPPTPGAQTPAQEE19 pKa = 4.41VIKK22 pKa = 10.27VTGQTPLTNDD32 pKa = 2.97QQGPYY37 pKa = 10.3LLGVAQKK44 pKa = 8.56ITAEE48 pKa = 4.12EE49 pKa = 4.21IEE51 pKa = 4.31RR52 pKa = 11.84SNAKK56 pKa = 8.41TLTQQLKK63 pKa = 8.29NTLVSVNINDD73 pKa = 3.59VQNNPFQPDD82 pKa = 3.13VQYY85 pKa = 11.16RR86 pKa = 11.84GFTASPLLGLPQGISVYY103 pKa = 10.87LDD105 pKa = 3.38GIRR108 pKa = 11.84FNEE111 pKa = 4.1PFGDD115 pKa = 3.78TVNWDD120 pKa = 4.91LIALTAIDD128 pKa = 4.72NVTLYY133 pKa = 10.75SGSNPVFGQNTLGGALALSVKK154 pKa = 10.71DD155 pKa = 3.22GFTYY159 pKa = 10.51DD160 pKa = 3.31GHH162 pKa = 6.28EE163 pKa = 4.57LSAQAGSFGEE173 pKa = 4.18KK174 pKa = 9.63QLNVQSGGTSGNWGYY189 pKa = 11.18YY190 pKa = 10.49LNLNHH195 pKa = 6.39YY196 pKa = 9.86EE197 pKa = 3.87EE198 pKa = 5.67DD199 pKa = 3.21GWRR202 pKa = 11.84DD203 pKa = 3.43GSPSEE208 pKa = 4.23VKK210 pKa = 10.41QYY212 pKa = 10.95LANLSYY218 pKa = 9.75KK219 pKa = 10.06TDD221 pKa = 4.07DD222 pKa = 3.6LTADD226 pKa = 4.43LLFATNNNTMIGNGTVPIEE245 pKa = 4.05LLEE248 pKa = 4.25QEE250 pKa = 4.7SSTAIYY256 pKa = 7.54TQPDD260 pKa = 3.38QTKK263 pKa = 9.68TNLNMLGLNITAALTEE279 pKa = 4.47DD280 pKa = 3.2SSLAVNAYY288 pKa = 8.37YY289 pKa = 10.49RR290 pKa = 11.84KK291 pKa = 10.31NEE293 pKa = 3.65ISSINGDD300 pKa = 3.36DD301 pKa = 3.65SDD303 pKa = 4.4YY304 pKa = 11.64DD305 pKa = 3.53EE306 pKa = 6.24CEE308 pKa = 3.89FGTAVTLCEE317 pKa = 5.32GEE319 pKa = 5.4DD320 pKa = 4.35DD321 pKa = 6.08DD322 pKa = 7.0DD323 pKa = 6.76DD324 pKa = 5.51EE325 pKa = 6.44IDD327 pKa = 4.48GIEE330 pKa = 4.15SHH332 pKa = 7.04GDD334 pKa = 3.26DD335 pKa = 4.33DD336 pKa = 6.32DD337 pKa = 4.51EE338 pKa = 6.59GEE340 pKa = 4.39EE341 pKa = 3.99IEE343 pKa = 4.29PVHH346 pKa = 5.71FVGYY350 pKa = 10.81NEE352 pKa = 4.11GQYY355 pKa = 11.08LSDD358 pKa = 4.41IMPALDD364 pKa = 4.94ADD366 pKa = 5.45DD367 pKa = 5.22IDD369 pKa = 3.87GTYY372 pKa = 8.99NTGATDD378 pKa = 3.48NEE380 pKa = 4.68SFGFSAQLVTQWGNGHH396 pKa = 5.99EE397 pKa = 4.34LIIGGGADD405 pKa = 3.17KK406 pKa = 11.26ADD408 pKa = 3.08ISFASDD414 pKa = 3.1TQFAILHH421 pKa = 6.48NDD423 pKa = 3.01TAEE426 pKa = 4.72DD427 pKa = 3.89DD428 pKa = 4.3RR429 pKa = 11.84SVTPIPGLFDD439 pKa = 3.27QEE441 pKa = 4.46STVRR445 pKa = 11.84LDD447 pKa = 3.6TEE449 pKa = 4.02TTQYY453 pKa = 11.42YY454 pKa = 9.84FYY456 pKa = 10.7LANRR460 pKa = 11.84FKK462 pKa = 10.86LASDD466 pKa = 3.85LHH468 pKa = 8.15LDD470 pKa = 3.15IAGRR474 pKa = 11.84YY475 pKa = 8.0HH476 pKa = 8.01DD477 pKa = 5.91SNILMNDD484 pKa = 4.39LIDD487 pKa = 4.86DD488 pKa = 4.54GEE490 pKa = 4.9GSLDD494 pKa = 3.57GDD496 pKa = 3.69HH497 pKa = 7.4DD498 pKa = 4.63FSRR501 pKa = 11.84FNPAIGLSYY510 pKa = 10.45QISPQLTAKK519 pKa = 10.63LSYY522 pKa = 10.38SEE524 pKa = 4.61SSRR527 pKa = 11.84NPSPAEE533 pKa = 3.73LSCADD538 pKa = 4.11EE539 pKa = 5.82DD540 pKa = 5.48DD541 pKa = 4.21PCKK544 pKa = 10.66LPNGFVSDD552 pKa = 4.38PPLKK556 pKa = 10.01QVVVNTVEE564 pKa = 4.64LSLLGKK570 pKa = 10.04DD571 pKa = 4.18DD572 pKa = 3.77NLNYY576 pKa = 10.67GVTFFSTQSQDD587 pKa = 4.97DD588 pKa = 4.39IIFQQAGEE596 pKa = 4.29HH597 pKa = 6.43PARR600 pKa = 11.84GYY602 pKa = 10.03FVNIDD607 pKa = 3.17EE608 pKa = 4.5TQRR611 pKa = 11.84RR612 pKa = 11.84GVEE615 pKa = 3.87LSASYY620 pKa = 10.25RR621 pKa = 11.84GEE623 pKa = 4.07SFDD626 pKa = 3.21ITSSYY631 pKa = 11.36NYY633 pKa = 10.42LDD635 pKa = 3.62ATFEE639 pKa = 4.54SPFVSFSPVNPLGPNRR655 pKa = 11.84QVSPGDD661 pKa = 3.8TIPGQPQHH669 pKa = 5.9QIKK672 pKa = 10.95LIGQYY677 pKa = 9.76QATEE681 pKa = 4.03SFDD684 pKa = 3.26IGSEE688 pKa = 3.8LSYY691 pKa = 11.2ASEE694 pKa = 4.02SYY696 pKa = 10.98YY697 pKa = 10.92RR698 pKa = 11.84GDD700 pKa = 3.39EE701 pKa = 4.12ANEE704 pKa = 3.7NQQIGSHH711 pKa = 4.7TVVNLFANYY720 pKa = 10.17QITQQLALEE729 pKa = 4.26LRR731 pKa = 11.84VDD733 pKa = 3.77NVFDD737 pKa = 3.55RR738 pKa = 11.84QYY740 pKa = 10.37FTFGAYY746 pKa = 10.04GEE748 pKa = 4.17ADD750 pKa = 3.05EE751 pKa = 5.0VLEE754 pKa = 4.08NVYY757 pKa = 10.63PDD759 pKa = 3.35IEE761 pKa = 4.38DD762 pKa = 4.08PYY764 pKa = 11.18FVGPAKK770 pKa = 9.99PRR772 pKa = 11.84SVSVEE777 pKa = 3.63LSYY780 pKa = 11.19QFF782 pKa = 4.51
Molecular weight: 85.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N0WMW9|A0A2N0WMW9_9GAMM Cysteine--tRNA ligase OS=Alteromonadales bacterium alter-6D02 OX=2058087 GN=cysS PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.35GFRR19 pKa = 11.84ARR21 pKa = 11.84SATKK25 pKa = 10.08GGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.49GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.35GFRR19 pKa = 11.84ARR21 pKa = 11.84SATKK25 pKa = 10.08GGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.49GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 4.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1160949 |
23 |
3442 |
340.0 |
37.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.321 ± 0.044 | 0.988 ± 0.013 |
5.558 ± 0.036 | 5.944 ± 0.041 |
4.187 ± 0.029 | 6.374 ± 0.04 |
2.304 ± 0.021 | 6.707 ± 0.034 |
5.446 ± 0.037 | 10.57 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.332 ± 0.019 | 4.62 ± 0.032 |
3.666 ± 0.029 | 5.282 ± 0.044 |
4.18 ± 0.033 | 7.003 ± 0.034 |
5.515 ± 0.042 | 6.807 ± 0.036 |
1.112 ± 0.014 | 3.083 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
