
Zhizhongheella caldifontis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Zhizhongheella
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
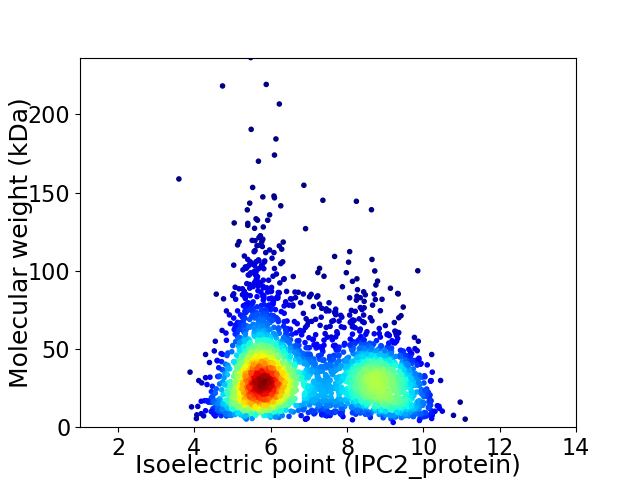
Virtual 2D-PAGE plot for 3372 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S5STL0|A0A2S5STL0_9BURK Helix-turn-helix transcriptional regulator OS=Zhizhongheella caldifontis OX=1452508 GN=C1704_10400 PE=4 SV=1
MM1 pKa = 7.53AGSYY5 pKa = 10.27TDD7 pKa = 3.12IAGNSGSAASLTLAADD23 pKa = 3.77IAAPVVDD30 pKa = 5.8LDD32 pKa = 3.97ASSPGTGYY40 pKa = 11.0VVTFTEE46 pKa = 4.09NGSPVSIGDD55 pKa = 3.37IDD57 pKa = 5.76LSITDD62 pKa = 4.34ADD64 pKa = 3.91STTLSGATITLSNPQADD81 pKa = 3.89DD82 pKa = 3.58VLAVGSMPAGITATVVGNVVTLSGTASLADD112 pKa = 3.54YY113 pKa = 8.88EE114 pKa = 4.23AALRR118 pKa = 11.84SISFEE123 pKa = 4.21NTSEE127 pKa = 4.72APDD130 pKa = 3.59TTPRR134 pKa = 11.84TISVSVVDD142 pKa = 4.37ANGNASAPAVATVQVVSVDD161 pKa = 3.75DD162 pKa = 4.73APVHH166 pKa = 6.06ALPGPVTMLEE176 pKa = 4.11DD177 pKa = 3.5SQIYY181 pKa = 9.26LLPGTYY187 pKa = 8.59VTDD190 pKa = 3.5IDD192 pKa = 4.39SDD194 pKa = 4.85LITTTVSVPVGTLVLGSTDD213 pKa = 3.06GVTVSGNGTGVVVLSGSPSAVNAALIWTTYY243 pKa = 9.54TPPQDD248 pKa = 3.59YY249 pKa = 10.96NGVVDD254 pKa = 3.68ITVTTTDD261 pKa = 3.07GTSTVTDD268 pKa = 3.86SLNVTITPVNDD279 pKa = 3.23APVAVDD285 pKa = 3.7DD286 pKa = 4.42TFSTAEE292 pKa = 4.09DD293 pKa = 3.56TAITITPAQLLANDD307 pKa = 3.93IDD309 pKa = 4.06VDD311 pKa = 3.96GDD313 pKa = 3.94ALSVATIHH321 pKa = 6.69NITNGSISIVGGNIVFTPNPNYY343 pKa = 10.24HH344 pKa = 6.39GPASFDD350 pKa = 3.43YY351 pKa = 10.14TISDD355 pKa = 3.3GRR357 pKa = 11.84GGFSTARR364 pKa = 11.84ATIDD368 pKa = 3.4VLSVDD373 pKa = 5.06DD374 pKa = 4.89LPEE377 pKa = 4.18TADD380 pKa = 3.57VNASGAEE387 pKa = 3.94DD388 pKa = 3.74SVIAIALSGSDD399 pKa = 3.37VDD401 pKa = 4.88NVITGYY407 pKa = 11.08VIGTLPANGVLYY419 pKa = 10.48RR420 pKa = 11.84DD421 pKa = 3.5AALTQVVSAGDD432 pKa = 3.69VVSEE436 pKa = 4.0TTLYY440 pKa = 10.18FRR442 pKa = 11.84PADD445 pKa = 3.3NWNGNTSFQYY455 pKa = 10.76AAQNDD460 pKa = 4.05AGISDD465 pKa = 4.27PTPATVVITVTPVNDD480 pKa = 3.37APVAVNDD487 pKa = 4.03SFTTAEE493 pKa = 4.3DD494 pKa = 3.52TAITITPAQLLANDD508 pKa = 3.95IDD510 pKa = 3.93VDD512 pKa = 4.14GDD514 pKa = 3.9VLSVATIHH522 pKa = 6.68NITNGSISIVGGNIVFTPNPNYY544 pKa = 10.24HH545 pKa = 6.39GPASFDD551 pKa = 3.44YY552 pKa = 10.34TISDD556 pKa = 3.74GQGGFSTATATISVTPVNDD575 pKa = 3.27APQAVDD581 pKa = 4.86DD582 pKa = 5.09GPSMLTGLRR591 pKa = 11.84GEE593 pKa = 4.15YY594 pKa = 9.27FAYY597 pKa = 10.23VEE599 pKa = 4.64GMSGPNLTSVQQVRR613 pKa = 11.84AFADD617 pKa = 3.44ANTADD622 pKa = 3.61AVFNATTLTYY632 pKa = 11.09NEE634 pKa = 4.19INGNLGADD642 pKa = 3.43GRR644 pKa = 11.84LQAFLGSDD652 pKa = 3.42AASLSVDD659 pKa = 3.9PPNSSDD665 pKa = 3.68AILRR669 pKa = 11.84FTGTLDD675 pKa = 3.7LAAGTYY681 pKa = 9.14ILRR684 pKa = 11.84VRR686 pKa = 11.84ADD688 pKa = 3.52DD689 pKa = 5.11GYY691 pKa = 11.57SIMIDD696 pKa = 3.24GVVVAEE702 pKa = 4.43VNANQAPTTTTHH714 pKa = 7.18AAFTLPTGGPHH725 pKa = 7.01SIDD728 pKa = 3.45IIYY731 pKa = 9.58WDD733 pKa = 3.41QGGQAVFGVEE743 pKa = 3.96LSADD747 pKa = 3.63NGSTFQPLGQYY758 pKa = 7.2TLRR761 pKa = 11.84HH762 pKa = 5.2PTYY765 pKa = 8.3TTAEE769 pKa = 4.12DD770 pKa = 3.58TALVISTATLLANDD784 pKa = 4.36TDD786 pKa = 3.86VDD788 pKa = 4.18GDD790 pKa = 3.98PLSVVSVQDD799 pKa = 3.55AVNGTVTYY807 pKa = 10.59SGGQIIFTPAPDD819 pKa = 3.62YY820 pKa = 10.96HH821 pKa = 7.93GPASFTYY828 pKa = 9.47TVSDD832 pKa = 4.1GNGGTSTATVSLNILSVNDD851 pKa = 3.68MPTTANVTATGAEE864 pKa = 4.13DD865 pKa = 3.46SVLAVTLAGADD876 pKa = 2.9IDD878 pKa = 4.12GTVTGFVIQNLPANGTLYY896 pKa = 11.15ADD898 pKa = 4.19AARR901 pKa = 11.84TQVISAGSVVPGPVVYY917 pKa = 9.02FQPSANWNGSTTFQYY932 pKa = 11.13AARR935 pKa = 11.84DD936 pKa = 3.91NEE938 pKa = 4.31GGTDD942 pKa = 3.7STPATATLTVTPVNDD957 pKa = 3.34APVNVVPGLQTFNEE971 pKa = 4.2DD972 pKa = 3.03TSRR975 pKa = 11.84VFSVANGNAISVADD989 pKa = 4.05IDD991 pKa = 4.39SASLTTSLSVSHH1003 pKa = 6.11GTLTLATTSGLTVSGNGTGTVTLSGSAAAINAALNGTTYY1042 pKa = 10.33TPAANVHH1049 pKa = 5.86GATTLTVTTSDD1060 pKa = 3.5GSLSTTSTVPMNVVAVADD1078 pKa = 4.24TPLLGAQEE1086 pKa = 4.98IIQVLNPGSTMISTGAGMPSNFTRR1110 pKa = 11.84GTNHH1114 pKa = 6.77GDD1116 pKa = 3.3GVSAANLEE1124 pKa = 4.38AEE1126 pKa = 4.73LGLAPGALSQFNPPGSHH1143 pKa = 6.76AGNVTAIDD1151 pKa = 4.07GKK1153 pKa = 9.68LTTANYY1159 pKa = 10.0SLSAGTTVNFNWSFLNGEE1177 pKa = 4.63DD1178 pKa = 3.89LDD1180 pKa = 4.37SEE1182 pKa = 4.15IRR1184 pKa = 11.84AGYY1187 pKa = 9.25NDD1189 pKa = 4.35LVILVVTDD1197 pKa = 3.58PAGNRR1202 pKa = 11.84QAIQVTSSEE1211 pKa = 4.13QAGASTGTLNGTYY1224 pKa = 10.08AYY1226 pKa = 7.69TAASAGSYY1234 pKa = 10.01KK1235 pKa = 10.18FDD1237 pKa = 2.96WVIVNGRR1244 pKa = 11.84DD1245 pKa = 3.12AGKK1248 pKa = 10.29DD1249 pKa = 3.35SRR1251 pKa = 11.84LHH1253 pKa = 5.28LTSTSFSTSGSAHH1266 pKa = 6.57GAPVALALFAALTDD1280 pKa = 3.85TDD1282 pKa = 3.91GSEE1285 pKa = 4.28SLSVTVAGVPSGARR1299 pKa = 11.84LSSGTNLGSGVWSLSPADD1317 pKa = 4.32LPGLQLLPASGYY1329 pKa = 9.32TGTINLNVTATATEE1343 pKa = 3.99ASNGSVASTSRR1354 pKa = 11.84SISITIAEE1362 pKa = 4.43TGNTVHH1368 pKa = 6.88SGSQSGDD1375 pKa = 3.65SVSGASGNSSDD1386 pKa = 5.7LIHH1389 pKa = 7.19GYY1391 pKa = 10.38AGNDD1395 pKa = 3.66TINAGNGHH1403 pKa = 6.67DD1404 pKa = 4.31LVHH1407 pKa = 6.84GGTGNDD1413 pKa = 3.55TLNGEE1418 pKa = 4.69DD1419 pKa = 3.92GHH1421 pKa = 7.1DD1422 pKa = 3.67VLYY1425 pKa = 11.15GGAGNDD1431 pKa = 3.89TLNGGAGNDD1440 pKa = 3.33ILVGGTGNDD1449 pKa = 3.52TLTGGTGADD1458 pKa = 3.26VFRR1461 pKa = 11.84WHH1463 pKa = 6.77FGDD1466 pKa = 3.79GGVVGTPAVDD1476 pKa = 4.24TITDD1480 pKa = 4.41FNNSTGPQGDD1490 pKa = 3.87VLDD1493 pKa = 4.97LRR1495 pKa = 11.84DD1496 pKa = 3.79LLVGEE1501 pKa = 4.31SHH1503 pKa = 6.9GNLSNYY1509 pKa = 10.03LHH1511 pKa = 7.13FSTSGSGASTTTTISISTTGGFAGGFSASAVDD1543 pKa = 5.68QVIQLNNVDD1552 pKa = 4.45LVQSFTSDD1560 pKa = 3.12QQIIQDD1566 pKa = 4.06LLARR1570 pKa = 11.84GKK1572 pKa = 10.41LLSDD1576 pKa = 3.94
MM1 pKa = 7.53AGSYY5 pKa = 10.27TDD7 pKa = 3.12IAGNSGSAASLTLAADD23 pKa = 3.77IAAPVVDD30 pKa = 5.8LDD32 pKa = 3.97ASSPGTGYY40 pKa = 11.0VVTFTEE46 pKa = 4.09NGSPVSIGDD55 pKa = 3.37IDD57 pKa = 5.76LSITDD62 pKa = 4.34ADD64 pKa = 3.91STTLSGATITLSNPQADD81 pKa = 3.89DD82 pKa = 3.58VLAVGSMPAGITATVVGNVVTLSGTASLADD112 pKa = 3.54YY113 pKa = 8.88EE114 pKa = 4.23AALRR118 pKa = 11.84SISFEE123 pKa = 4.21NTSEE127 pKa = 4.72APDD130 pKa = 3.59TTPRR134 pKa = 11.84TISVSVVDD142 pKa = 4.37ANGNASAPAVATVQVVSVDD161 pKa = 3.75DD162 pKa = 4.73APVHH166 pKa = 6.06ALPGPVTMLEE176 pKa = 4.11DD177 pKa = 3.5SQIYY181 pKa = 9.26LLPGTYY187 pKa = 8.59VTDD190 pKa = 3.5IDD192 pKa = 4.39SDD194 pKa = 4.85LITTTVSVPVGTLVLGSTDD213 pKa = 3.06GVTVSGNGTGVVVLSGSPSAVNAALIWTTYY243 pKa = 9.54TPPQDD248 pKa = 3.59YY249 pKa = 10.96NGVVDD254 pKa = 3.68ITVTTTDD261 pKa = 3.07GTSTVTDD268 pKa = 3.86SLNVTITPVNDD279 pKa = 3.23APVAVDD285 pKa = 3.7DD286 pKa = 4.42TFSTAEE292 pKa = 4.09DD293 pKa = 3.56TAITITPAQLLANDD307 pKa = 3.93IDD309 pKa = 4.06VDD311 pKa = 3.96GDD313 pKa = 3.94ALSVATIHH321 pKa = 6.69NITNGSISIVGGNIVFTPNPNYY343 pKa = 10.24HH344 pKa = 6.39GPASFDD350 pKa = 3.43YY351 pKa = 10.14TISDD355 pKa = 3.3GRR357 pKa = 11.84GGFSTARR364 pKa = 11.84ATIDD368 pKa = 3.4VLSVDD373 pKa = 5.06DD374 pKa = 4.89LPEE377 pKa = 4.18TADD380 pKa = 3.57VNASGAEE387 pKa = 3.94DD388 pKa = 3.74SVIAIALSGSDD399 pKa = 3.37VDD401 pKa = 4.88NVITGYY407 pKa = 11.08VIGTLPANGVLYY419 pKa = 10.48RR420 pKa = 11.84DD421 pKa = 3.5AALTQVVSAGDD432 pKa = 3.69VVSEE436 pKa = 4.0TTLYY440 pKa = 10.18FRR442 pKa = 11.84PADD445 pKa = 3.3NWNGNTSFQYY455 pKa = 10.76AAQNDD460 pKa = 4.05AGISDD465 pKa = 4.27PTPATVVITVTPVNDD480 pKa = 3.37APVAVNDD487 pKa = 4.03SFTTAEE493 pKa = 4.3DD494 pKa = 3.52TAITITPAQLLANDD508 pKa = 3.95IDD510 pKa = 3.93VDD512 pKa = 4.14GDD514 pKa = 3.9VLSVATIHH522 pKa = 6.68NITNGSISIVGGNIVFTPNPNYY544 pKa = 10.24HH545 pKa = 6.39GPASFDD551 pKa = 3.44YY552 pKa = 10.34TISDD556 pKa = 3.74GQGGFSTATATISVTPVNDD575 pKa = 3.27APQAVDD581 pKa = 4.86DD582 pKa = 5.09GPSMLTGLRR591 pKa = 11.84GEE593 pKa = 4.15YY594 pKa = 9.27FAYY597 pKa = 10.23VEE599 pKa = 4.64GMSGPNLTSVQQVRR613 pKa = 11.84AFADD617 pKa = 3.44ANTADD622 pKa = 3.61AVFNATTLTYY632 pKa = 11.09NEE634 pKa = 4.19INGNLGADD642 pKa = 3.43GRR644 pKa = 11.84LQAFLGSDD652 pKa = 3.42AASLSVDD659 pKa = 3.9PPNSSDD665 pKa = 3.68AILRR669 pKa = 11.84FTGTLDD675 pKa = 3.7LAAGTYY681 pKa = 9.14ILRR684 pKa = 11.84VRR686 pKa = 11.84ADD688 pKa = 3.52DD689 pKa = 5.11GYY691 pKa = 11.57SIMIDD696 pKa = 3.24GVVVAEE702 pKa = 4.43VNANQAPTTTTHH714 pKa = 7.18AAFTLPTGGPHH725 pKa = 7.01SIDD728 pKa = 3.45IIYY731 pKa = 9.58WDD733 pKa = 3.41QGGQAVFGVEE743 pKa = 3.96LSADD747 pKa = 3.63NGSTFQPLGQYY758 pKa = 7.2TLRR761 pKa = 11.84HH762 pKa = 5.2PTYY765 pKa = 8.3TTAEE769 pKa = 4.12DD770 pKa = 3.58TALVISTATLLANDD784 pKa = 4.36TDD786 pKa = 3.86VDD788 pKa = 4.18GDD790 pKa = 3.98PLSVVSVQDD799 pKa = 3.55AVNGTVTYY807 pKa = 10.59SGGQIIFTPAPDD819 pKa = 3.62YY820 pKa = 10.96HH821 pKa = 7.93GPASFTYY828 pKa = 9.47TVSDD832 pKa = 4.1GNGGTSTATVSLNILSVNDD851 pKa = 3.68MPTTANVTATGAEE864 pKa = 4.13DD865 pKa = 3.46SVLAVTLAGADD876 pKa = 2.9IDD878 pKa = 4.12GTVTGFVIQNLPANGTLYY896 pKa = 11.15ADD898 pKa = 4.19AARR901 pKa = 11.84TQVISAGSVVPGPVVYY917 pKa = 9.02FQPSANWNGSTTFQYY932 pKa = 11.13AARR935 pKa = 11.84DD936 pKa = 3.91NEE938 pKa = 4.31GGTDD942 pKa = 3.7STPATATLTVTPVNDD957 pKa = 3.34APVNVVPGLQTFNEE971 pKa = 4.2DD972 pKa = 3.03TSRR975 pKa = 11.84VFSVANGNAISVADD989 pKa = 4.05IDD991 pKa = 4.39SASLTTSLSVSHH1003 pKa = 6.11GTLTLATTSGLTVSGNGTGTVTLSGSAAAINAALNGTTYY1042 pKa = 10.33TPAANVHH1049 pKa = 5.86GATTLTVTTSDD1060 pKa = 3.5GSLSTTSTVPMNVVAVADD1078 pKa = 4.24TPLLGAQEE1086 pKa = 4.98IIQVLNPGSTMISTGAGMPSNFTRR1110 pKa = 11.84GTNHH1114 pKa = 6.77GDD1116 pKa = 3.3GVSAANLEE1124 pKa = 4.38AEE1126 pKa = 4.73LGLAPGALSQFNPPGSHH1143 pKa = 6.76AGNVTAIDD1151 pKa = 4.07GKK1153 pKa = 9.68LTTANYY1159 pKa = 10.0SLSAGTTVNFNWSFLNGEE1177 pKa = 4.63DD1178 pKa = 3.89LDD1180 pKa = 4.37SEE1182 pKa = 4.15IRR1184 pKa = 11.84AGYY1187 pKa = 9.25NDD1189 pKa = 4.35LVILVVTDD1197 pKa = 3.58PAGNRR1202 pKa = 11.84QAIQVTSSEE1211 pKa = 4.13QAGASTGTLNGTYY1224 pKa = 10.08AYY1226 pKa = 7.69TAASAGSYY1234 pKa = 10.01KK1235 pKa = 10.18FDD1237 pKa = 2.96WVIVNGRR1244 pKa = 11.84DD1245 pKa = 3.12AGKK1248 pKa = 10.29DD1249 pKa = 3.35SRR1251 pKa = 11.84LHH1253 pKa = 5.28LTSTSFSTSGSAHH1266 pKa = 6.57GAPVALALFAALTDD1280 pKa = 3.85TDD1282 pKa = 3.91GSEE1285 pKa = 4.28SLSVTVAGVPSGARR1299 pKa = 11.84LSSGTNLGSGVWSLSPADD1317 pKa = 4.32LPGLQLLPASGYY1329 pKa = 9.32TGTINLNVTATATEE1343 pKa = 3.99ASNGSVASTSRR1354 pKa = 11.84SISITIAEE1362 pKa = 4.43TGNTVHH1368 pKa = 6.88SGSQSGDD1375 pKa = 3.65SVSGASGNSSDD1386 pKa = 5.7LIHH1389 pKa = 7.19GYY1391 pKa = 10.38AGNDD1395 pKa = 3.66TINAGNGHH1403 pKa = 6.67DD1404 pKa = 4.31LVHH1407 pKa = 6.84GGTGNDD1413 pKa = 3.55TLNGEE1418 pKa = 4.69DD1419 pKa = 3.92GHH1421 pKa = 7.1DD1422 pKa = 3.67VLYY1425 pKa = 11.15GGAGNDD1431 pKa = 3.89TLNGGAGNDD1440 pKa = 3.33ILVGGTGNDD1449 pKa = 3.52TLTGGTGADD1458 pKa = 3.26VFRR1461 pKa = 11.84WHH1463 pKa = 6.77FGDD1466 pKa = 3.79GGVVGTPAVDD1476 pKa = 4.24TITDD1480 pKa = 4.41FNNSTGPQGDD1490 pKa = 3.87VLDD1493 pKa = 4.97LRR1495 pKa = 11.84DD1496 pKa = 3.79LLVGEE1501 pKa = 4.31SHH1503 pKa = 6.9GNLSNYY1509 pKa = 10.03LHH1511 pKa = 7.13FSTSGSGASTTTTISISTTGGFAGGFSASAVDD1543 pKa = 5.68QVIQLNNVDD1552 pKa = 4.45LVQSFTSDD1560 pKa = 3.12QQIIQDD1566 pKa = 4.06LLARR1570 pKa = 11.84GKK1572 pKa = 10.41LLSDD1576 pKa = 3.94
Molecular weight: 158.71 kDa
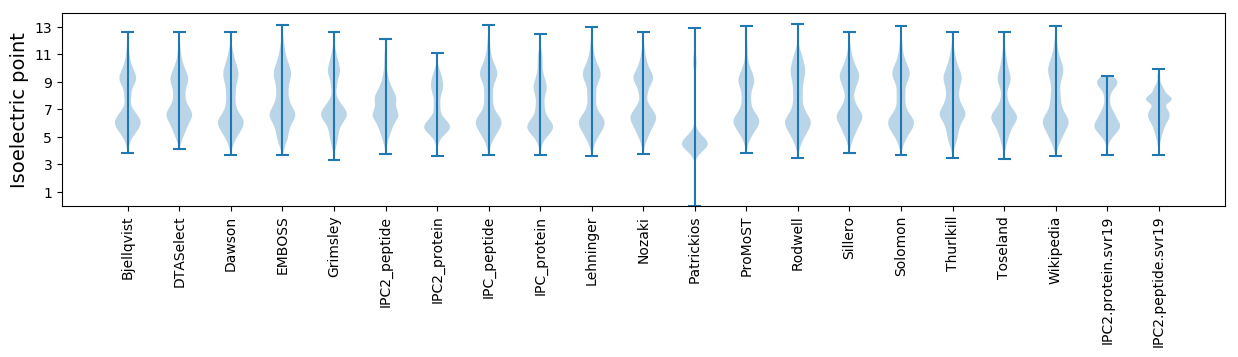
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S5SXU2|A0A2S5SXU2_9BURK NAD-dependent succinate-semialdehyde dehydrogenase OS=Zhizhongheella caldifontis OX=1452508 GN=C1704_04290 PE=4 SV=1
MM1 pKa = 7.19KK2 pKa = 10.25RR3 pKa = 11.84SYY5 pKa = 10.63QPSKK9 pKa = 9.66VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.67TKK25 pKa = 10.48GGRR28 pKa = 11.84KK29 pKa = 9.03VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.19KK2 pKa = 10.25RR3 pKa = 11.84SYY5 pKa = 10.63QPSKK9 pKa = 9.66VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.67TKK25 pKa = 10.48GGRR28 pKa = 11.84KK29 pKa = 9.03VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1112190 |
28 |
2190 |
329.8 |
35.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.84 ± 0.057 | 0.893 ± 0.014 |
5.2 ± 0.027 | 5.618 ± 0.037 |
3.372 ± 0.026 | 8.281 ± 0.034 |
2.458 ± 0.025 | 3.977 ± 0.035 |
2.756 ± 0.035 | 11.19 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.344 ± 0.019 | 2.209 ± 0.022 |
5.527 ± 0.031 | 3.998 ± 0.024 |
7.781 ± 0.04 | 4.918 ± 0.026 |
4.951 ± 0.028 | 7.928 ± 0.032 |
1.624 ± 0.023 | 2.132 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
