
Raphanus sativus cryptic virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
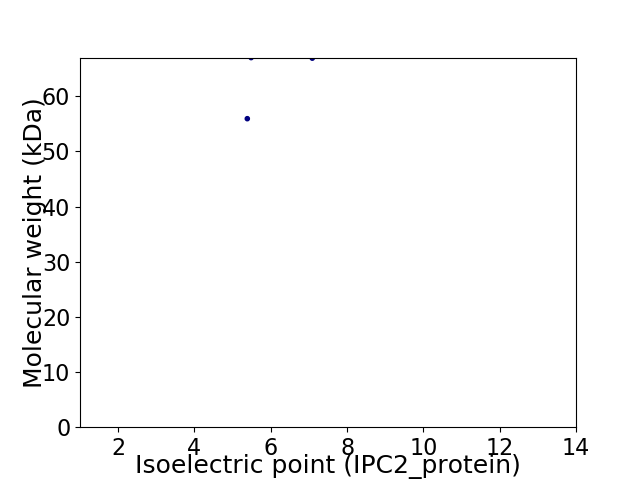
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q52UL1|Q52UL1_9VIRU RNA-dependent RNA polymerase OS=Raphanus sativus cryptic virus 1 OX=348449 PE=4 SV=2
MM1 pKa = 7.69AHH3 pKa = 6.35RR4 pKa = 11.84TPTNAPALPQVDD16 pKa = 5.17GINPNLPPNDD26 pKa = 4.32PGTVAAAPNARR37 pKa = 11.84NLHH40 pKa = 5.71LEE42 pKa = 4.1RR43 pKa = 11.84EE44 pKa = 4.21QEE46 pKa = 3.88ARR48 pKa = 11.84QDD50 pKa = 2.98RR51 pKa = 11.84AAAAFVPNTRR61 pKa = 11.84FAVRR65 pKa = 11.84RR66 pKa = 11.84PTIRR70 pKa = 11.84PSPAHH75 pKa = 6.12SAPDD79 pKa = 3.52LEE81 pKa = 4.54TALFDD86 pKa = 4.06SAVNHH91 pKa = 6.79PPTPVSYY98 pKa = 9.83PSTSSYY104 pKa = 10.68IPNFTSAFYY113 pKa = 10.71YY114 pKa = 9.65LNKK117 pKa = 9.34MDD119 pKa = 4.32SLMVQTLNWTNNCSGWVPPYY139 pKa = 8.97SQIYY143 pKa = 10.19ISMLLYY149 pKa = 9.74LQVMRR154 pKa = 11.84AMKK157 pKa = 10.17KK158 pKa = 10.51AGVLRR163 pKa = 11.84PNSEE167 pKa = 3.92LSHH170 pKa = 6.63LFNEE174 pKa = 4.71MSTIFPFEE182 pKa = 4.13SLMVPGPLVNLFEE195 pKa = 5.63NITAFRR201 pKa = 11.84PLQTDD206 pKa = 2.97SFGNVTPFLPAEE218 pKa = 4.63PGWSNATFFAPNGSLVRR235 pKa = 11.84HH236 pKa = 6.49LPHH239 pKa = 7.04IPALISRR246 pKa = 11.84LRR248 pKa = 11.84RR249 pKa = 11.84ICEE252 pKa = 3.81TASEE256 pKa = 4.26NGLNDD261 pKa = 3.08ISFSAHH267 pKa = 3.93HH268 pKa = 6.84HH269 pKa = 5.35GPEE272 pKa = 4.63FISEE276 pKa = 4.28LFGHH280 pKa = 6.79ICDD283 pKa = 4.2NDD285 pKa = 3.76LPEE288 pKa = 4.32QLLLLTPGLATSYY301 pKa = 11.03SGTLYY306 pKa = 10.71LWRR309 pKa = 11.84QARR312 pKa = 11.84SQLQRR317 pKa = 11.84SLFPEE322 pKa = 4.32ALTVNDD328 pKa = 3.95VVPNTWTSFLCLDD341 pKa = 4.78NDD343 pKa = 4.54DD344 pKa = 4.7SWFSPLAAMMNKK356 pKa = 9.09YY357 pKa = 8.98CQYY360 pKa = 8.41WHH362 pKa = 7.02GSAPLSSIPADD373 pKa = 3.67GSAAGSMICNEE384 pKa = 4.43LNGSSIYY391 pKa = 10.61RR392 pKa = 11.84LAQWTAPIMDD402 pKa = 4.7PPADD406 pKa = 3.75EE407 pKa = 5.15DD408 pKa = 4.04APPVIAEE415 pKa = 3.89PGYY418 pKa = 10.69YY419 pKa = 9.57YY420 pKa = 9.89PNPNPVLIFDD430 pKa = 4.33ARR432 pKa = 11.84TCIEE436 pKa = 5.3DD437 pKa = 3.05ISSAHH442 pKa = 6.48MFSAMTFHH450 pKa = 7.54PNLIPYY456 pKa = 7.76GANRR460 pKa = 11.84ANFLKK465 pKa = 10.75GQFWNCHH472 pKa = 5.33PPSYY476 pKa = 10.08SAPAHH481 pKa = 4.59QVYY484 pKa = 8.85PAVSALIARR493 pKa = 11.84EE494 pKa = 3.92YY495 pKa = 10.76HH496 pKa = 6.41SEE498 pKa = 4.25TNLSSDD504 pKa = 3.33KK505 pKa = 11.34
MM1 pKa = 7.69AHH3 pKa = 6.35RR4 pKa = 11.84TPTNAPALPQVDD16 pKa = 5.17GINPNLPPNDD26 pKa = 4.32PGTVAAAPNARR37 pKa = 11.84NLHH40 pKa = 5.71LEE42 pKa = 4.1RR43 pKa = 11.84EE44 pKa = 4.21QEE46 pKa = 3.88ARR48 pKa = 11.84QDD50 pKa = 2.98RR51 pKa = 11.84AAAAFVPNTRR61 pKa = 11.84FAVRR65 pKa = 11.84RR66 pKa = 11.84PTIRR70 pKa = 11.84PSPAHH75 pKa = 6.12SAPDD79 pKa = 3.52LEE81 pKa = 4.54TALFDD86 pKa = 4.06SAVNHH91 pKa = 6.79PPTPVSYY98 pKa = 9.83PSTSSYY104 pKa = 10.68IPNFTSAFYY113 pKa = 10.71YY114 pKa = 9.65LNKK117 pKa = 9.34MDD119 pKa = 4.32SLMVQTLNWTNNCSGWVPPYY139 pKa = 8.97SQIYY143 pKa = 10.19ISMLLYY149 pKa = 9.74LQVMRR154 pKa = 11.84AMKK157 pKa = 10.17KK158 pKa = 10.51AGVLRR163 pKa = 11.84PNSEE167 pKa = 3.92LSHH170 pKa = 6.63LFNEE174 pKa = 4.71MSTIFPFEE182 pKa = 4.13SLMVPGPLVNLFEE195 pKa = 5.63NITAFRR201 pKa = 11.84PLQTDD206 pKa = 2.97SFGNVTPFLPAEE218 pKa = 4.63PGWSNATFFAPNGSLVRR235 pKa = 11.84HH236 pKa = 6.49LPHH239 pKa = 7.04IPALISRR246 pKa = 11.84LRR248 pKa = 11.84RR249 pKa = 11.84ICEE252 pKa = 3.81TASEE256 pKa = 4.26NGLNDD261 pKa = 3.08ISFSAHH267 pKa = 3.93HH268 pKa = 6.84HH269 pKa = 5.35GPEE272 pKa = 4.63FISEE276 pKa = 4.28LFGHH280 pKa = 6.79ICDD283 pKa = 4.2NDD285 pKa = 3.76LPEE288 pKa = 4.32QLLLLTPGLATSYY301 pKa = 11.03SGTLYY306 pKa = 10.71LWRR309 pKa = 11.84QARR312 pKa = 11.84SQLQRR317 pKa = 11.84SLFPEE322 pKa = 4.32ALTVNDD328 pKa = 3.95VVPNTWTSFLCLDD341 pKa = 4.78NDD343 pKa = 4.54DD344 pKa = 4.7SWFSPLAAMMNKK356 pKa = 9.09YY357 pKa = 8.98CQYY360 pKa = 8.41WHH362 pKa = 7.02GSAPLSSIPADD373 pKa = 3.67GSAAGSMICNEE384 pKa = 4.43LNGSSIYY391 pKa = 10.61RR392 pKa = 11.84LAQWTAPIMDD402 pKa = 4.7PPADD406 pKa = 3.75EE407 pKa = 5.15DD408 pKa = 4.04APPVIAEE415 pKa = 3.89PGYY418 pKa = 10.69YY419 pKa = 9.57YY420 pKa = 9.89PNPNPVLIFDD430 pKa = 4.33ARR432 pKa = 11.84TCIEE436 pKa = 5.3DD437 pKa = 3.05ISSAHH442 pKa = 6.48MFSAMTFHH450 pKa = 7.54PNLIPYY456 pKa = 7.76GANRR460 pKa = 11.84ANFLKK465 pKa = 10.75GQFWNCHH472 pKa = 5.33PPSYY476 pKa = 10.08SAPAHH481 pKa = 4.59QVYY484 pKa = 8.85PAVSALIARR493 pKa = 11.84EE494 pKa = 3.92YY495 pKa = 10.76HH496 pKa = 6.41SEE498 pKa = 4.25TNLSSDD504 pKa = 3.33KK505 pKa = 11.34
Molecular weight: 55.94 kDa
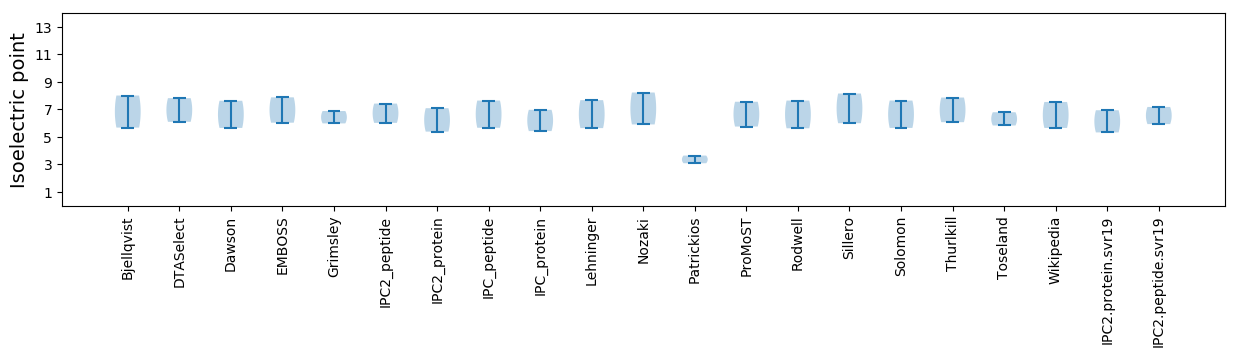
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q52UL1|Q52UL1_9VIRU RNA-dependent RNA polymerase OS=Raphanus sativus cryptic virus 1 OX=348449 PE=4 SV=2
MM1 pKa = 7.77SNLEE5 pKa = 3.96YY6 pKa = 10.79LGLDD10 pKa = 3.51HH11 pKa = 7.33HH12 pKa = 6.96WPAIPRR18 pKa = 11.84FAKK21 pKa = 10.55SPNDD25 pKa = 3.16WYY27 pKa = 10.56FACQQLVRR35 pKa = 11.84SSICLYY41 pKa = 10.94SSLLGSNDD49 pKa = 3.03TDD51 pKa = 3.75TVLNGYY57 pKa = 8.48YY58 pKa = 9.9RR59 pKa = 11.84SHH61 pKa = 7.9ADD63 pKa = 3.29EE64 pKa = 4.63DD65 pKa = 3.94TAEE68 pKa = 4.12QFFMRR73 pKa = 11.84YY74 pKa = 9.22DD75 pKa = 3.49VEE77 pKa = 4.22PFDD80 pKa = 5.19IIKK83 pKa = 10.72DD84 pKa = 3.69NYY86 pKa = 9.61YY87 pKa = 10.46SQAFDD92 pKa = 3.88TVTEE96 pKa = 4.4WFRR99 pKa = 11.84PSAPIHH105 pKa = 5.83PVHH108 pKa = 5.95FTDD111 pKa = 4.69VRR113 pKa = 11.84WYY115 pKa = 8.09PWKK118 pKa = 10.35ISTSAEE124 pKa = 3.67RR125 pKa = 11.84PFTHH129 pKa = 7.86DD130 pKa = 3.63PLLKK134 pKa = 10.59KK135 pKa = 10.17KK136 pKa = 10.36VQLSKK141 pKa = 11.11QLGLLDD147 pKa = 3.59NARR150 pKa = 11.84MSFHH154 pKa = 6.63NCYY157 pKa = 10.64NDD159 pKa = 2.49IFTYY163 pKa = 9.9CRR165 pKa = 11.84HH166 pKa = 5.16YY167 pKa = 9.06THH169 pKa = 6.74EE170 pKa = 4.48VKK172 pKa = 10.5DD173 pKa = 3.77ARR175 pKa = 11.84PVTLHH180 pKa = 7.02HH181 pKa = 7.16IDD183 pKa = 3.78LHH185 pKa = 6.06VKK187 pKa = 8.97PALVRR192 pKa = 11.84SGEE195 pKa = 4.01PPKK198 pKa = 10.33IRR200 pKa = 11.84TVFGVPKK207 pKa = 10.45SLIFAEE213 pKa = 5.95AMFFWPLFSDD223 pKa = 3.91YY224 pKa = 9.05FTNSEE229 pKa = 4.67TPLLWNYY236 pKa = 7.32EE237 pKa = 4.18TLNGGWYY244 pKa = 10.1RR245 pKa = 11.84LNDD248 pKa = 3.31EE249 pKa = 6.3FYY251 pKa = 10.64QQWQSFCTIFNLDD264 pKa = 2.84WSEE267 pKa = 3.84FDD269 pKa = 3.13MRR271 pKa = 11.84VYY273 pKa = 10.71FSMLDD278 pKa = 3.5DD279 pKa = 4.53CRR281 pKa = 11.84DD282 pKa = 3.35AVKK285 pKa = 10.5SYY287 pKa = 11.11FCFCGNYY294 pKa = 9.8CPTRR298 pKa = 11.84TYY300 pKa = 7.53PTCRR304 pKa = 11.84TNPQRR309 pKa = 11.84LQNLWNWIGTAYY321 pKa = 10.01KK322 pKa = 8.88DD323 pKa = 4.1TPCTTTTGKK332 pKa = 10.2VYY334 pKa = 10.48RR335 pKa = 11.84RR336 pKa = 11.84RR337 pKa = 11.84FAGMPSGIFCTQFWDD352 pKa = 3.81SFYY355 pKa = 10.73NCIMVVTTLEE365 pKa = 3.98ALGFRR370 pKa = 11.84ITDD373 pKa = 3.7RR374 pKa = 11.84YY375 pKa = 8.65FLKK378 pKa = 11.0VLGDD382 pKa = 3.75DD383 pKa = 4.1VIFGILKK390 pKa = 9.73HH391 pKa = 6.87IPISKK396 pKa = 9.29WADD399 pKa = 3.23FLQDD403 pKa = 3.48FSTEE407 pKa = 3.16ARR409 pKa = 11.84RR410 pKa = 11.84RR411 pKa = 11.84FNSKK415 pKa = 10.43LNSKK419 pKa = 9.94KK420 pKa = 10.5CGASSGIHH428 pKa = 5.55GAQVLSYY435 pKa = 9.9INWNGYY441 pKa = 7.42PKK443 pKa = 10.0RR444 pKa = 11.84DD445 pKa = 3.79SNQLLAQLLHH455 pKa = 6.58PKK457 pKa = 9.59SLRR460 pKa = 11.84DD461 pKa = 3.42TYY463 pKa = 10.58PRR465 pKa = 11.84LMARR469 pKa = 11.84AIGIYY474 pKa = 9.5YY475 pKa = 10.06ASCGDD480 pKa = 3.59PKK482 pKa = 10.44IRR484 pKa = 11.84PICNHH489 pKa = 6.58IYY491 pKa = 10.67SEE493 pKa = 4.1LKK495 pKa = 10.13YY496 pKa = 10.89AGFTPSSTGLHH507 pKa = 6.18GLFDD511 pKa = 5.16PNASIGFIEE520 pKa = 5.15LDD522 pKa = 3.46HH523 pKa = 6.72FPSEE527 pKa = 4.37NEE529 pKa = 4.03VTCRR533 pKa = 11.84LHH535 pKa = 7.03RR536 pKa = 11.84KK537 pKa = 8.4SKK539 pKa = 10.67RR540 pKa = 11.84SAEE543 pKa = 4.01LQALYY548 pKa = 9.73WPRR551 pKa = 11.84DD552 pKa = 3.48HH553 pKa = 7.15FLEE556 pKa = 4.45EE557 pKa = 4.48AGSSRR562 pKa = 11.84NCPLSFQVEE571 pKa = 4.79TII573 pKa = 3.51
MM1 pKa = 7.77SNLEE5 pKa = 3.96YY6 pKa = 10.79LGLDD10 pKa = 3.51HH11 pKa = 7.33HH12 pKa = 6.96WPAIPRR18 pKa = 11.84FAKK21 pKa = 10.55SPNDD25 pKa = 3.16WYY27 pKa = 10.56FACQQLVRR35 pKa = 11.84SSICLYY41 pKa = 10.94SSLLGSNDD49 pKa = 3.03TDD51 pKa = 3.75TVLNGYY57 pKa = 8.48YY58 pKa = 9.9RR59 pKa = 11.84SHH61 pKa = 7.9ADD63 pKa = 3.29EE64 pKa = 4.63DD65 pKa = 3.94TAEE68 pKa = 4.12QFFMRR73 pKa = 11.84YY74 pKa = 9.22DD75 pKa = 3.49VEE77 pKa = 4.22PFDD80 pKa = 5.19IIKK83 pKa = 10.72DD84 pKa = 3.69NYY86 pKa = 9.61YY87 pKa = 10.46SQAFDD92 pKa = 3.88TVTEE96 pKa = 4.4WFRR99 pKa = 11.84PSAPIHH105 pKa = 5.83PVHH108 pKa = 5.95FTDD111 pKa = 4.69VRR113 pKa = 11.84WYY115 pKa = 8.09PWKK118 pKa = 10.35ISTSAEE124 pKa = 3.67RR125 pKa = 11.84PFTHH129 pKa = 7.86DD130 pKa = 3.63PLLKK134 pKa = 10.59KK135 pKa = 10.17KK136 pKa = 10.36VQLSKK141 pKa = 11.11QLGLLDD147 pKa = 3.59NARR150 pKa = 11.84MSFHH154 pKa = 6.63NCYY157 pKa = 10.64NDD159 pKa = 2.49IFTYY163 pKa = 9.9CRR165 pKa = 11.84HH166 pKa = 5.16YY167 pKa = 9.06THH169 pKa = 6.74EE170 pKa = 4.48VKK172 pKa = 10.5DD173 pKa = 3.77ARR175 pKa = 11.84PVTLHH180 pKa = 7.02HH181 pKa = 7.16IDD183 pKa = 3.78LHH185 pKa = 6.06VKK187 pKa = 8.97PALVRR192 pKa = 11.84SGEE195 pKa = 4.01PPKK198 pKa = 10.33IRR200 pKa = 11.84TVFGVPKK207 pKa = 10.45SLIFAEE213 pKa = 5.95AMFFWPLFSDD223 pKa = 3.91YY224 pKa = 9.05FTNSEE229 pKa = 4.67TPLLWNYY236 pKa = 7.32EE237 pKa = 4.18TLNGGWYY244 pKa = 10.1RR245 pKa = 11.84LNDD248 pKa = 3.31EE249 pKa = 6.3FYY251 pKa = 10.64QQWQSFCTIFNLDD264 pKa = 2.84WSEE267 pKa = 3.84FDD269 pKa = 3.13MRR271 pKa = 11.84VYY273 pKa = 10.71FSMLDD278 pKa = 3.5DD279 pKa = 4.53CRR281 pKa = 11.84DD282 pKa = 3.35AVKK285 pKa = 10.5SYY287 pKa = 11.11FCFCGNYY294 pKa = 9.8CPTRR298 pKa = 11.84TYY300 pKa = 7.53PTCRR304 pKa = 11.84TNPQRR309 pKa = 11.84LQNLWNWIGTAYY321 pKa = 10.01KK322 pKa = 8.88DD323 pKa = 4.1TPCTTTTGKK332 pKa = 10.2VYY334 pKa = 10.48RR335 pKa = 11.84RR336 pKa = 11.84RR337 pKa = 11.84FAGMPSGIFCTQFWDD352 pKa = 3.81SFYY355 pKa = 10.73NCIMVVTTLEE365 pKa = 3.98ALGFRR370 pKa = 11.84ITDD373 pKa = 3.7RR374 pKa = 11.84YY375 pKa = 8.65FLKK378 pKa = 11.0VLGDD382 pKa = 3.75DD383 pKa = 4.1VIFGILKK390 pKa = 9.73HH391 pKa = 6.87IPISKK396 pKa = 9.29WADD399 pKa = 3.23FLQDD403 pKa = 3.48FSTEE407 pKa = 3.16ARR409 pKa = 11.84RR410 pKa = 11.84RR411 pKa = 11.84FNSKK415 pKa = 10.43LNSKK419 pKa = 9.94KK420 pKa = 10.5CGASSGIHH428 pKa = 5.55GAQVLSYY435 pKa = 9.9INWNGYY441 pKa = 7.42PKK443 pKa = 10.0RR444 pKa = 11.84DD445 pKa = 3.79SNQLLAQLLHH455 pKa = 6.58PKK457 pKa = 9.59SLRR460 pKa = 11.84DD461 pKa = 3.42TYY463 pKa = 10.58PRR465 pKa = 11.84LMARR469 pKa = 11.84AIGIYY474 pKa = 9.5YY475 pKa = 10.06ASCGDD480 pKa = 3.59PKK482 pKa = 10.44IRR484 pKa = 11.84PICNHH489 pKa = 6.58IYY491 pKa = 10.67SEE493 pKa = 4.1LKK495 pKa = 10.13YY496 pKa = 10.89AGFTPSSTGLHH507 pKa = 6.18GLFDD511 pKa = 5.16PNASIGFIEE520 pKa = 5.15LDD522 pKa = 3.46HH523 pKa = 6.72FPSEE527 pKa = 4.37NEE529 pKa = 4.03VTCRR533 pKa = 11.84LHH535 pKa = 7.03RR536 pKa = 11.84KK537 pKa = 8.4SKK539 pKa = 10.67RR540 pKa = 11.84SAEE543 pKa = 4.01LQALYY548 pKa = 9.73WPRR551 pKa = 11.84DD552 pKa = 3.48HH553 pKa = 7.15FLEE556 pKa = 4.45EE557 pKa = 4.48AGSSRR562 pKa = 11.84NCPLSFQVEE571 pKa = 4.79TII573 pKa = 3.51
Molecular weight: 66.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1078 |
505 |
573 |
539.0 |
61.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.514 ± 1.716 | 2.412 ± 0.549 |
5.288 ± 0.749 | 4.082 ± 0.051 |
6.03 ± 0.716 | 4.36 ± 0.265 |
3.432 ± 0.044 | 4.824 ± 0.179 |
2.968 ± 1.182 | 9.091 ± 0.275 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.134 ± 0.424 | 5.844 ± 0.853 |
8.071 ± 1.741 | 3.061 ± 0.06 |
5.473 ± 0.61 | 8.442 ± 0.574 |
5.844 ± 0.33 | 4.082 ± 0.051 |
2.319 ± 0.356 | 4.731 ± 0.643 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
