
Aliicoccus persicus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Staphylococcaceae; Aliicoccus
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
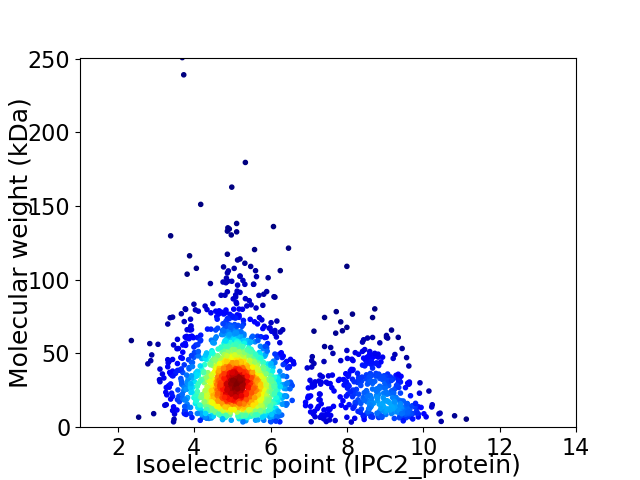
Virtual 2D-PAGE plot for 1983 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A662Z3Y1|A0A662Z3Y1_9STAP Uncharacterized protein OS=Aliicoccus persicus OX=930138 GN=SAMN05192557_0616 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.67KK3 pKa = 9.42MIKK6 pKa = 10.25LFTVSMLIGTTVAYY20 pKa = 10.4FGTTTHH26 pKa = 6.5AQDD29 pKa = 3.34EE30 pKa = 4.66ADD32 pKa = 3.51TVEE35 pKa = 3.91ISMTIGNDD43 pKa = 2.89IDD45 pKa = 4.16SLDD48 pKa = 3.95PFLAQATLTDD58 pKa = 3.69SVIYY62 pKa = 10.6NIFEE66 pKa = 4.28GLLDD70 pKa = 4.18AEE72 pKa = 4.82TDD74 pKa = 3.73GSLVPHH80 pKa = 7.26LAHH83 pKa = 7.48DD84 pKa = 4.16YY85 pKa = 10.33EE86 pKa = 5.59ISDD89 pKa = 4.58DD90 pKa = 3.68ALTYY94 pKa = 10.59TFFLEE99 pKa = 4.39EE100 pKa = 4.32GVTFHH105 pKa = 7.57DD106 pKa = 4.92GSDD109 pKa = 3.69FTAEE113 pKa = 4.07DD114 pKa = 3.31VVYY117 pKa = 9.27TYY119 pKa = 11.52SKK121 pKa = 10.82LAGLDD126 pKa = 3.77GEE128 pKa = 5.4DD129 pKa = 3.59ALSDD133 pKa = 2.95KK134 pKa = 10.77WEE136 pKa = 4.08VVEE139 pKa = 5.35SVEE142 pKa = 4.29ATDD145 pKa = 4.79DD146 pKa = 3.73YY147 pKa = 11.05TVEE150 pKa = 4.31VTLSNPDD157 pKa = 3.11SGFLARR163 pKa = 11.84TIRR166 pKa = 11.84AIVPADD172 pKa = 3.81YY173 pKa = 10.54DD174 pKa = 3.68DD175 pKa = 5.13HH176 pKa = 6.33ATNPVGTGPYY186 pKa = 9.34TFVEE190 pKa = 4.15HH191 pKa = 7.36SIDD194 pKa = 4.11QRR196 pKa = 11.84VTLEE200 pKa = 3.98KK201 pKa = 10.46FDD203 pKa = 6.78DD204 pKa = 4.0YY205 pKa = 11.61FQGNDD210 pKa = 3.03FDD212 pKa = 4.79VDD214 pKa = 3.94VVNFDD219 pKa = 3.62VMPDD223 pKa = 3.32EE224 pKa = 4.46EE225 pKa = 4.44TALLAFQAGEE235 pKa = 4.0VDD237 pKa = 5.33FITAVSDD244 pKa = 3.6QNLSRR249 pKa = 11.84VPEE252 pKa = 4.34DD253 pKa = 3.47ANVVSGPLNMVMMLAMNHH271 pKa = 5.2EE272 pKa = 4.53VEE274 pKa = 4.63PLDD277 pKa = 3.85SVEE280 pKa = 4.34VRR282 pKa = 11.84QAMNMVVDD290 pKa = 3.97KK291 pKa = 11.59QEE293 pKa = 4.48VIDD296 pKa = 4.4VAIDD300 pKa = 3.67GNGDD304 pKa = 3.67ALHH307 pKa = 6.44TFMSPAMDD315 pKa = 4.35FYY317 pKa = 11.49FNEE320 pKa = 4.23EE321 pKa = 3.85LVDD324 pKa = 3.98YY325 pKa = 7.96YY326 pKa = 11.14TVDD329 pKa = 3.15VEE331 pKa = 4.62TAQSLMAEE339 pKa = 4.43AGYY342 pKa = 11.03EE343 pKa = 3.99DD344 pKa = 4.46GFSIEE349 pKa = 4.16MKK351 pKa = 10.45VPNNAQLYY359 pKa = 9.01IDD361 pKa = 4.61AAQVLQAQLALINIDD376 pKa = 3.41LTLDD380 pKa = 3.62VIEE383 pKa = 4.61FSTWLEE389 pKa = 3.64DD390 pKa = 3.6VYY392 pKa = 11.4NEE394 pKa = 4.61RR395 pKa = 11.84EE396 pKa = 4.07FEE398 pKa = 4.1STIIGFTGKK407 pKa = 10.24LDD409 pKa = 4.22PYY411 pKa = 11.1DD412 pKa = 3.82VMIRR416 pKa = 11.84MEE418 pKa = 3.94TGYY421 pKa = 10.89AYY423 pKa = 11.29NFMNVDD429 pKa = 2.94IEE431 pKa = 4.81GYY433 pKa = 10.5NEE435 pKa = 3.86ALEE438 pKa = 4.39AAISAPSPEE447 pKa = 4.34EE448 pKa = 3.64SATLYY453 pKa = 10.91KK454 pKa = 10.18EE455 pKa = 4.14AQEE458 pKa = 4.59LLTEE462 pKa = 3.92NAAAVFLMDD471 pKa = 4.32PARR474 pKa = 11.84NSVMRR479 pKa = 11.84SGISGFEE486 pKa = 3.62MYY488 pKa = 9.97PYY490 pKa = 10.43EE491 pKa = 4.92RR492 pKa = 11.84YY493 pKa = 9.26VVKK496 pKa = 10.81DD497 pKa = 3.41LVVEE501 pKa = 4.45
MM1 pKa = 7.59KK2 pKa = 10.67KK3 pKa = 9.42MIKK6 pKa = 10.25LFTVSMLIGTTVAYY20 pKa = 10.4FGTTTHH26 pKa = 6.5AQDD29 pKa = 3.34EE30 pKa = 4.66ADD32 pKa = 3.51TVEE35 pKa = 3.91ISMTIGNDD43 pKa = 2.89IDD45 pKa = 4.16SLDD48 pKa = 3.95PFLAQATLTDD58 pKa = 3.69SVIYY62 pKa = 10.6NIFEE66 pKa = 4.28GLLDD70 pKa = 4.18AEE72 pKa = 4.82TDD74 pKa = 3.73GSLVPHH80 pKa = 7.26LAHH83 pKa = 7.48DD84 pKa = 4.16YY85 pKa = 10.33EE86 pKa = 5.59ISDD89 pKa = 4.58DD90 pKa = 3.68ALTYY94 pKa = 10.59TFFLEE99 pKa = 4.39EE100 pKa = 4.32GVTFHH105 pKa = 7.57DD106 pKa = 4.92GSDD109 pKa = 3.69FTAEE113 pKa = 4.07DD114 pKa = 3.31VVYY117 pKa = 9.27TYY119 pKa = 11.52SKK121 pKa = 10.82LAGLDD126 pKa = 3.77GEE128 pKa = 5.4DD129 pKa = 3.59ALSDD133 pKa = 2.95KK134 pKa = 10.77WEE136 pKa = 4.08VVEE139 pKa = 5.35SVEE142 pKa = 4.29ATDD145 pKa = 4.79DD146 pKa = 3.73YY147 pKa = 11.05TVEE150 pKa = 4.31VTLSNPDD157 pKa = 3.11SGFLARR163 pKa = 11.84TIRR166 pKa = 11.84AIVPADD172 pKa = 3.81YY173 pKa = 10.54DD174 pKa = 3.68DD175 pKa = 5.13HH176 pKa = 6.33ATNPVGTGPYY186 pKa = 9.34TFVEE190 pKa = 4.15HH191 pKa = 7.36SIDD194 pKa = 4.11QRR196 pKa = 11.84VTLEE200 pKa = 3.98KK201 pKa = 10.46FDD203 pKa = 6.78DD204 pKa = 4.0YY205 pKa = 11.61FQGNDD210 pKa = 3.03FDD212 pKa = 4.79VDD214 pKa = 3.94VVNFDD219 pKa = 3.62VMPDD223 pKa = 3.32EE224 pKa = 4.46EE225 pKa = 4.44TALLAFQAGEE235 pKa = 4.0VDD237 pKa = 5.33FITAVSDD244 pKa = 3.6QNLSRR249 pKa = 11.84VPEE252 pKa = 4.34DD253 pKa = 3.47ANVVSGPLNMVMMLAMNHH271 pKa = 5.2EE272 pKa = 4.53VEE274 pKa = 4.63PLDD277 pKa = 3.85SVEE280 pKa = 4.34VRR282 pKa = 11.84QAMNMVVDD290 pKa = 3.97KK291 pKa = 11.59QEE293 pKa = 4.48VIDD296 pKa = 4.4VAIDD300 pKa = 3.67GNGDD304 pKa = 3.67ALHH307 pKa = 6.44TFMSPAMDD315 pKa = 4.35FYY317 pKa = 11.49FNEE320 pKa = 4.23EE321 pKa = 3.85LVDD324 pKa = 3.98YY325 pKa = 7.96YY326 pKa = 11.14TVDD329 pKa = 3.15VEE331 pKa = 4.62TAQSLMAEE339 pKa = 4.43AGYY342 pKa = 11.03EE343 pKa = 3.99DD344 pKa = 4.46GFSIEE349 pKa = 4.16MKK351 pKa = 10.45VPNNAQLYY359 pKa = 9.01IDD361 pKa = 4.61AAQVLQAQLALINIDD376 pKa = 3.41LTLDD380 pKa = 3.62VIEE383 pKa = 4.61FSTWLEE389 pKa = 3.64DD390 pKa = 3.6VYY392 pKa = 11.4NEE394 pKa = 4.61RR395 pKa = 11.84EE396 pKa = 4.07FEE398 pKa = 4.1STIIGFTGKK407 pKa = 10.24LDD409 pKa = 4.22PYY411 pKa = 11.1DD412 pKa = 3.82VMIRR416 pKa = 11.84MEE418 pKa = 3.94TGYY421 pKa = 10.89AYY423 pKa = 11.29NFMNVDD429 pKa = 2.94IEE431 pKa = 4.81GYY433 pKa = 10.5NEE435 pKa = 3.86ALEE438 pKa = 4.39AAISAPSPEE447 pKa = 4.34EE448 pKa = 3.64SATLYY453 pKa = 10.91KK454 pKa = 10.18EE455 pKa = 4.14AQEE458 pKa = 4.59LLTEE462 pKa = 3.92NAAAVFLMDD471 pKa = 4.32PARR474 pKa = 11.84NSVMRR479 pKa = 11.84SGISGFEE486 pKa = 3.62MYY488 pKa = 9.97PYY490 pKa = 10.43EE491 pKa = 4.92RR492 pKa = 11.84YY493 pKa = 9.26VVKK496 pKa = 10.81DD497 pKa = 3.41LVVEE501 pKa = 4.45
Molecular weight: 55.75 kDa
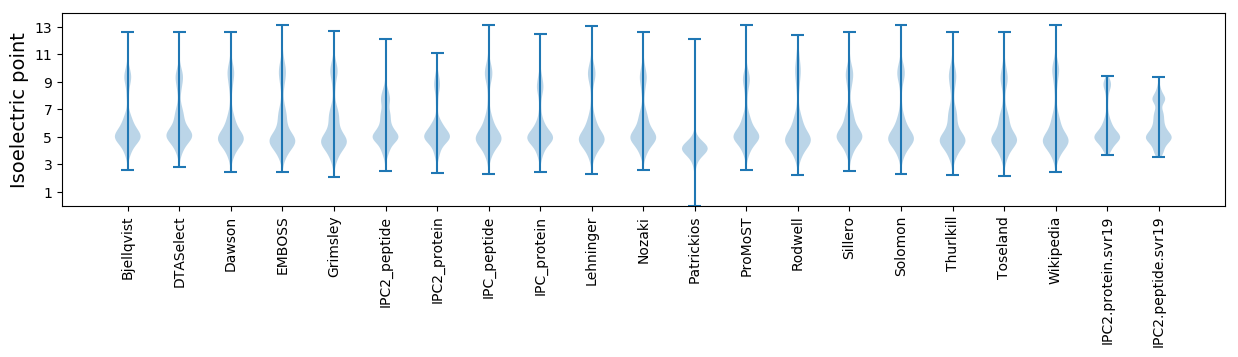
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A662Z256|A0A662Z256_9STAP Transcriptional regulator LacI family OS=Aliicoccus persicus OX=930138 GN=SAMN05192557_0664 PE=4 SV=1
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.35QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.87VHH17 pKa = 5.77GFRR20 pKa = 11.84KK21 pKa = 10.02RR22 pKa = 11.84MSTKK26 pKa = 10.16NGRR29 pKa = 11.84HH30 pKa = 3.62VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 8.94VLSAA45 pKa = 4.11
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.35QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.87VHH17 pKa = 5.77GFRR20 pKa = 11.84KK21 pKa = 10.02RR22 pKa = 11.84MSTKK26 pKa = 10.16NGRR29 pKa = 11.84HH30 pKa = 3.62VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 8.94VLSAA45 pKa = 4.11
Molecular weight: 5.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
597384 |
30 |
2252 |
301.3 |
34.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.311 ± 0.066 | 0.493 ± 0.015 |
6.541 ± 0.069 | 8.158 ± 0.074 |
4.602 ± 0.047 | 6.271 ± 0.048 |
2.218 ± 0.029 | 8.152 ± 0.057 |
6.022 ± 0.06 | 9.216 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.9 ± 0.026 | 5.012 ± 0.042 |
3.182 ± 0.026 | 3.207 ± 0.029 |
4.126 ± 0.041 | 6.021 ± 0.037 |
5.639 ± 0.043 | 7.351 ± 0.044 |
0.732 ± 0.017 | 3.847 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
