
Oleomonas sp. K1W22B-8
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Oleomonas; unclassified Oleomonas
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
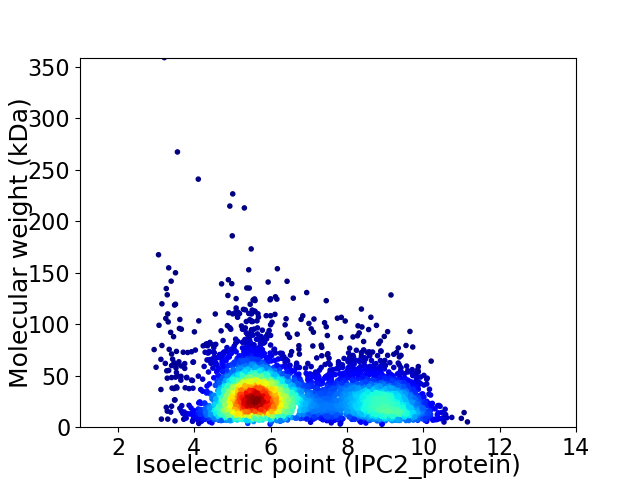
Virtual 2D-PAGE plot for 5024 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A418WDE3|A0A418WDE3_9PROT DNA topoisomerase 4 subunit A OS=Oleomonas sp. K1W22B-8 OX=2320859 GN=parC PE=3 SV=1
MM1 pKa = 7.63PAFAEE6 pKa = 4.39TFDD9 pKa = 5.02LADD12 pKa = 4.33LDD14 pKa = 4.44GEE16 pKa = 4.5NGFHH20 pKa = 6.52LVSEE24 pKa = 4.43AGSYY28 pKa = 10.19NFGYY32 pKa = 10.48SIATLGDD39 pKa = 3.49VNGDD43 pKa = 3.61GFDD46 pKa = 5.12DD47 pKa = 3.61IAVNSFDD54 pKa = 4.81GPTILLGGQGPFAPSTTVSNEE75 pKa = 3.65FPFEE79 pKa = 4.03TRR81 pKa = 11.84TATLPDD87 pKa = 4.53DD88 pKa = 3.68LTEE91 pKa = 4.16YY92 pKa = 10.44EE93 pKa = 4.23SWIVGIGDD101 pKa = 3.5VNGDD105 pKa = 3.33GFADD109 pKa = 4.07FDD111 pKa = 3.61HH112 pKa = 6.8HH113 pKa = 7.21AFYY116 pKa = 9.18ATEE119 pKa = 4.02TYY121 pKa = 10.64YY122 pKa = 10.61EE123 pKa = 4.13YY124 pKa = 10.46EE125 pKa = 4.47YY126 pKa = 11.1GDD128 pKa = 3.31SWTEE132 pKa = 3.96IFFHH136 pKa = 7.48DD137 pKa = 4.4YY138 pKa = 11.31GRR140 pKa = 11.84IQYY143 pKa = 10.26GGGSAATTGKK153 pKa = 10.44RR154 pKa = 11.84FGDD157 pKa = 3.56RR158 pKa = 11.84GLEE161 pKa = 4.06DD162 pKa = 4.68VDD164 pKa = 4.47PSWLVPLGDD173 pKa = 3.75VNGDD177 pKa = 3.42GFDD180 pKa = 3.74DD181 pKa = 3.89VVVYY185 pKa = 10.66VQDD188 pKa = 3.82GSHH191 pKa = 6.71PKK193 pKa = 10.13VVFGSATGLPDD204 pKa = 3.29NVNRR208 pKa = 11.84GSAPSLAFTGASPFEE223 pKa = 4.29INGAGDD229 pKa = 3.52VNGDD233 pKa = 4.0GITDD237 pKa = 3.69VVVGVSDD244 pKa = 3.52SSGVDD249 pKa = 2.95HH250 pKa = 6.11YY251 pKa = 11.63VVYY254 pKa = 10.61GGAGLAPGFFDD265 pKa = 4.64LTSLDD270 pKa = 3.39GTNGFKK276 pKa = 10.26LTDD279 pKa = 3.41ARR281 pKa = 11.84TVGTVGDD288 pKa = 3.79VDD290 pKa = 4.99GDD292 pKa = 4.01GLDD295 pKa = 4.0DD296 pKa = 4.78LVVSMSGRR304 pKa = 11.84TSSDD308 pKa = 2.7PYY310 pKa = 8.83YY311 pKa = 10.84VVYY314 pKa = 10.63GRR316 pKa = 11.84AAGPAAIDD324 pKa = 3.54ASAVDD329 pKa = 4.37GSDD332 pKa = 2.85GFQIVNAGTRR342 pKa = 11.84WVGRR346 pKa = 11.84AGDD349 pKa = 3.77INADD353 pKa = 3.63GLDD356 pKa = 4.16DD357 pKa = 4.54FLIGNDD363 pKa = 3.26YY364 pKa = 11.32VVFGRR369 pKa = 11.84ADD371 pKa = 3.21VGTGGLDD378 pKa = 3.19ATSVDD383 pKa = 3.97GDD385 pKa = 3.5NGFQIVATGIDD396 pKa = 3.87TTSSWGGLIVQAAGDD411 pKa = 3.93VNGDD415 pKa = 3.57GVDD418 pKa = 3.8DD419 pKa = 4.56LVAGTRR425 pKa = 11.84LEE427 pKa = 4.17NSVHH431 pKa = 6.23VILGHH436 pKa = 6.78AASAVNWVGTSARR449 pKa = 11.84EE450 pKa = 3.9NHH452 pKa = 6.48GGSPEE457 pKa = 3.97NDD459 pKa = 3.45VLNGAGGNDD468 pKa = 4.13YY469 pKa = 11.13LRR471 pKa = 11.84GWAVTTCC478 pKa = 3.65
MM1 pKa = 7.63PAFAEE6 pKa = 4.39TFDD9 pKa = 5.02LADD12 pKa = 4.33LDD14 pKa = 4.44GEE16 pKa = 4.5NGFHH20 pKa = 6.52LVSEE24 pKa = 4.43AGSYY28 pKa = 10.19NFGYY32 pKa = 10.48SIATLGDD39 pKa = 3.49VNGDD43 pKa = 3.61GFDD46 pKa = 5.12DD47 pKa = 3.61IAVNSFDD54 pKa = 4.81GPTILLGGQGPFAPSTTVSNEE75 pKa = 3.65FPFEE79 pKa = 4.03TRR81 pKa = 11.84TATLPDD87 pKa = 4.53DD88 pKa = 3.68LTEE91 pKa = 4.16YY92 pKa = 10.44EE93 pKa = 4.23SWIVGIGDD101 pKa = 3.5VNGDD105 pKa = 3.33GFADD109 pKa = 4.07FDD111 pKa = 3.61HH112 pKa = 6.8HH113 pKa = 7.21AFYY116 pKa = 9.18ATEE119 pKa = 4.02TYY121 pKa = 10.64YY122 pKa = 10.61EE123 pKa = 4.13YY124 pKa = 10.46EE125 pKa = 4.47YY126 pKa = 11.1GDD128 pKa = 3.31SWTEE132 pKa = 3.96IFFHH136 pKa = 7.48DD137 pKa = 4.4YY138 pKa = 11.31GRR140 pKa = 11.84IQYY143 pKa = 10.26GGGSAATTGKK153 pKa = 10.44RR154 pKa = 11.84FGDD157 pKa = 3.56RR158 pKa = 11.84GLEE161 pKa = 4.06DD162 pKa = 4.68VDD164 pKa = 4.47PSWLVPLGDD173 pKa = 3.75VNGDD177 pKa = 3.42GFDD180 pKa = 3.74DD181 pKa = 3.89VVVYY185 pKa = 10.66VQDD188 pKa = 3.82GSHH191 pKa = 6.71PKK193 pKa = 10.13VVFGSATGLPDD204 pKa = 3.29NVNRR208 pKa = 11.84GSAPSLAFTGASPFEE223 pKa = 4.29INGAGDD229 pKa = 3.52VNGDD233 pKa = 4.0GITDD237 pKa = 3.69VVVGVSDD244 pKa = 3.52SSGVDD249 pKa = 2.95HH250 pKa = 6.11YY251 pKa = 11.63VVYY254 pKa = 10.61GGAGLAPGFFDD265 pKa = 4.64LTSLDD270 pKa = 3.39GTNGFKK276 pKa = 10.26LTDD279 pKa = 3.41ARR281 pKa = 11.84TVGTVGDD288 pKa = 3.79VDD290 pKa = 4.99GDD292 pKa = 4.01GLDD295 pKa = 4.0DD296 pKa = 4.78LVVSMSGRR304 pKa = 11.84TSSDD308 pKa = 2.7PYY310 pKa = 8.83YY311 pKa = 10.84VVYY314 pKa = 10.63GRR316 pKa = 11.84AAGPAAIDD324 pKa = 3.54ASAVDD329 pKa = 4.37GSDD332 pKa = 2.85GFQIVNAGTRR342 pKa = 11.84WVGRR346 pKa = 11.84AGDD349 pKa = 3.77INADD353 pKa = 3.63GLDD356 pKa = 4.16DD357 pKa = 4.54FLIGNDD363 pKa = 3.26YY364 pKa = 11.32VVFGRR369 pKa = 11.84ADD371 pKa = 3.21VGTGGLDD378 pKa = 3.19ATSVDD383 pKa = 3.97GDD385 pKa = 3.5NGFQIVATGIDD396 pKa = 3.87TTSSWGGLIVQAAGDD411 pKa = 3.93VNGDD415 pKa = 3.57GVDD418 pKa = 3.8DD419 pKa = 4.56LVAGTRR425 pKa = 11.84LEE427 pKa = 4.17NSVHH431 pKa = 6.23VILGHH436 pKa = 6.78AASAVNWVGTSARR449 pKa = 11.84EE450 pKa = 3.9NHH452 pKa = 6.48GGSPEE457 pKa = 3.97NDD459 pKa = 3.45VLNGAGGNDD468 pKa = 4.13YY469 pKa = 11.13LRR471 pKa = 11.84GWAVTTCC478 pKa = 3.65
Molecular weight: 49.45 kDa
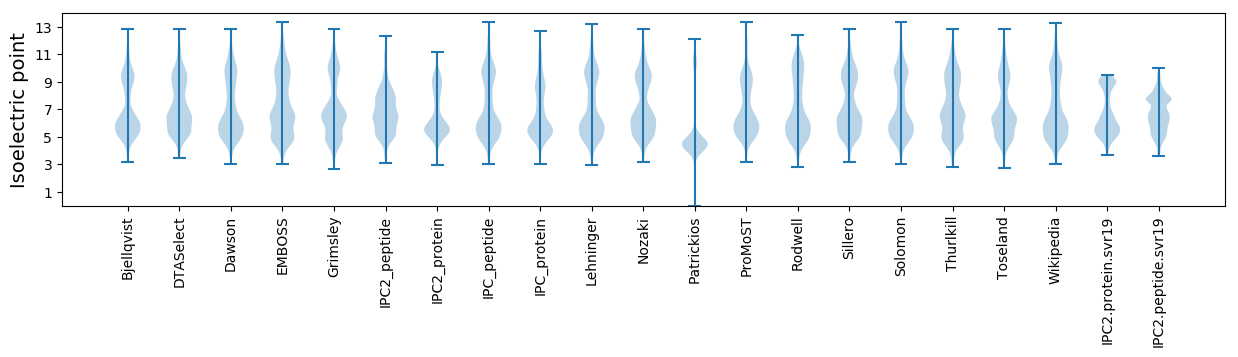
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A418WTK9|A0A418WTK9_9PROT Uncharacterized protein OS=Oleomonas sp. K1W22B-8 OX=2320859 GN=D3874_01865 PE=4 SV=1
MM1 pKa = 7.93PSPSSRR7 pKa = 11.84KK8 pKa = 6.83PWRR11 pKa = 11.84WRR13 pKa = 11.84LSWRR17 pKa = 11.84KK18 pKa = 9.38SPPRR22 pKa = 11.84KK23 pKa = 9.29PPPRR27 pKa = 11.84PSRR30 pKa = 11.84RR31 pKa = 11.84PLTAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84SPVVNRR45 pKa = 11.84RR46 pKa = 11.84PKK48 pKa = 9.9VVRR51 pKa = 11.84PPPMPGAANRR61 pKa = 11.84FSVANRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84SNGGPRR75 pKa = 11.84WRR77 pKa = 11.84SVGRR81 pKa = 11.84RR82 pKa = 11.84GPAANSNRR90 pKa = 11.84FNRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84GLQAGDD102 pKa = 3.69PWVSPAGRR110 pKa = 11.84CRR112 pKa = 11.84HH113 pKa = 5.21KK114 pKa = 11.07SMRR117 pKa = 11.84KK118 pKa = 9.49DD119 pKa = 3.76FPRR122 pKa = 5.34
MM1 pKa = 7.93PSPSSRR7 pKa = 11.84KK8 pKa = 6.83PWRR11 pKa = 11.84WRR13 pKa = 11.84LSWRR17 pKa = 11.84KK18 pKa = 9.38SPPRR22 pKa = 11.84KK23 pKa = 9.29PPPRR27 pKa = 11.84PSRR30 pKa = 11.84RR31 pKa = 11.84PLTAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84SPVVNRR45 pKa = 11.84RR46 pKa = 11.84PKK48 pKa = 9.9VVRR51 pKa = 11.84PPPMPGAANRR61 pKa = 11.84FSVANRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84SNGGPRR75 pKa = 11.84WRR77 pKa = 11.84SVGRR81 pKa = 11.84RR82 pKa = 11.84GPAANSNRR90 pKa = 11.84FNRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84GLQAGDD102 pKa = 3.69PWVSPAGRR110 pKa = 11.84CRR112 pKa = 11.84HH113 pKa = 5.21KK114 pKa = 11.07SMRR117 pKa = 11.84KK118 pKa = 9.49DD119 pKa = 3.76FPRR122 pKa = 5.34
Molecular weight: 14.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1531126 |
26 |
3535 |
304.8 |
32.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.388 ± 0.057 | 0.821 ± 0.011 |
5.925 ± 0.039 | 5.126 ± 0.032 |
3.647 ± 0.023 | 9.284 ± 0.055 |
1.984 ± 0.02 | 4.948 ± 0.024 |
2.906 ± 0.03 | 10.369 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.26 ± 0.02 | 2.421 ± 0.025 |
5.437 ± 0.031 | 2.975 ± 0.017 |
7.206 ± 0.04 | 4.836 ± 0.028 |
5.356 ± 0.028 | 7.56 ± 0.03 |
1.357 ± 0.015 | 2.194 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
