
Haliea sp.
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Haliea; unclassified Haliea
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
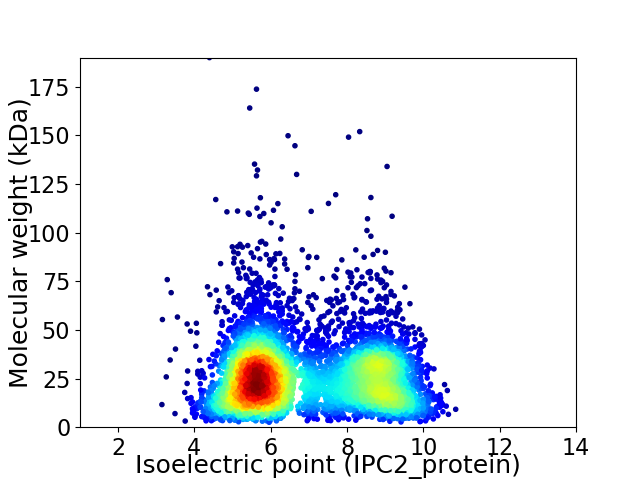
Virtual 2D-PAGE plot for 5874 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A519IFT1|A0A519IFT1_9GAMM VOC family protein OS=Haliea sp. OX=1932666 GN=EOO54_21010 PE=4 SV=1
MM1 pKa = 8.0ALMCPRR7 pKa = 11.84CGADD11 pKa = 3.04NGDD14 pKa = 3.44TARR17 pKa = 11.84FCKK20 pKa = 10.57SCGEE24 pKa = 4.21GLQAAAPVVQSSGTAPTEE42 pKa = 3.7PSAAPVCSSCGAVNSLGARR61 pKa = 11.84FCKK64 pKa = 10.52SCGTLLPAAASTAAAAAVVTPLPVTSPLQASGAAAFSPASPSLFTEE110 pKa = 5.1PPADD114 pKa = 3.7PVSTALPSPVAADD127 pKa = 3.89TSHH130 pKa = 6.3SPLWILLIAALLVLALGGGGYY151 pKa = 9.68WYY153 pKa = 10.21VQNRR157 pKa = 11.84GMPAFGALMGDD168 pKa = 3.69SDD170 pKa = 4.38PAAGSSDD177 pKa = 3.07AAEE180 pKa = 4.05VEE182 pKa = 4.26KK183 pKa = 11.01QEE185 pKa = 4.89EE186 pKa = 4.67VLTDD190 pKa = 3.26TAEE193 pKa = 4.21AADD196 pKa = 4.01EE197 pKa = 4.09EE198 pKa = 5.3VITEE202 pKa = 4.22VEE204 pKa = 3.62VDD206 pKa = 3.53AAADD210 pKa = 3.76TAADD214 pKa = 3.69AAADD218 pKa = 3.52AAAPAEE224 pKa = 4.57TQASPAA230 pKa = 3.69
MM1 pKa = 8.0ALMCPRR7 pKa = 11.84CGADD11 pKa = 3.04NGDD14 pKa = 3.44TARR17 pKa = 11.84FCKK20 pKa = 10.57SCGEE24 pKa = 4.21GLQAAAPVVQSSGTAPTEE42 pKa = 3.7PSAAPVCSSCGAVNSLGARR61 pKa = 11.84FCKK64 pKa = 10.52SCGTLLPAAASTAAAAAVVTPLPVTSPLQASGAAAFSPASPSLFTEE110 pKa = 5.1PPADD114 pKa = 3.7PVSTALPSPVAADD127 pKa = 3.89TSHH130 pKa = 6.3SPLWILLIAALLVLALGGGGYY151 pKa = 9.68WYY153 pKa = 10.21VQNRR157 pKa = 11.84GMPAFGALMGDD168 pKa = 3.69SDD170 pKa = 4.38PAAGSSDD177 pKa = 3.07AAEE180 pKa = 4.05VEE182 pKa = 4.26KK183 pKa = 11.01QEE185 pKa = 4.89EE186 pKa = 4.67VLTDD190 pKa = 3.26TAEE193 pKa = 4.21AADD196 pKa = 4.01EE197 pKa = 4.09EE198 pKa = 5.3VITEE202 pKa = 4.22VEE204 pKa = 3.62VDD206 pKa = 3.53AAADD210 pKa = 3.76TAADD214 pKa = 3.69AAADD218 pKa = 3.52AAAPAEE224 pKa = 4.57TQASPAA230 pKa = 3.69
Molecular weight: 22.48 kDa
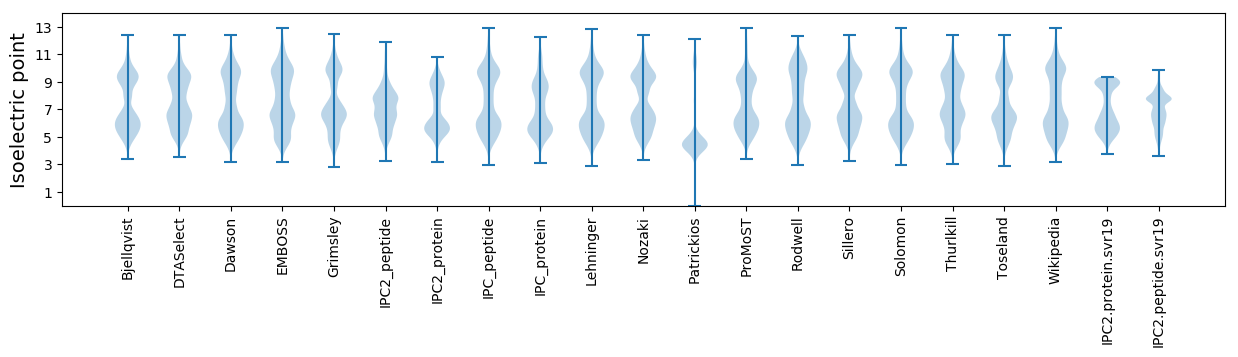
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A519IE07|A0A519IE07_9GAMM AAA family ATPase (Fragment) OS=Haliea sp. OX=1932666 GN=EOO54_21355 PE=4 SV=1
MM1 pKa = 7.52NLRR4 pKa = 11.84HH5 pKa = 5.81MEE7 pKa = 3.9VFRR10 pKa = 11.84AVMLTGGVVGAAEE23 pKa = 4.88LLHH26 pKa = 6.64ISQPAVSKK34 pKa = 10.73ILAQAQRR41 pKa = 11.84QLGFALFVRR50 pKa = 11.84IKK52 pKa = 10.81GRR54 pKa = 11.84LVPTAEE60 pKa = 4.15GQALHH65 pKa = 7.01AEE67 pKa = 4.56VEE69 pKa = 4.58TLWRR73 pKa = 11.84SVQRR77 pKa = 11.84VQATPRR83 pKa = 11.84SGTLTLAVSASLAPYY98 pKa = 9.74LVPRR102 pKa = 11.84TLATLARR109 pKa = 11.84RR110 pKa = 11.84FPGLQTRR117 pKa = 11.84MEE119 pKa = 4.17ILVTPIMVDD128 pKa = 3.0ALLDD132 pKa = 3.5RR133 pKa = 11.84TADD136 pKa = 3.05IGVAILPNDD145 pKa = 3.92HH146 pKa = 7.28PNLVQVSRR154 pKa = 11.84HH155 pKa = 5.79DD156 pKa = 3.57CGFACVLPEE165 pKa = 3.8GHH167 pKa = 7.08KK168 pKa = 10.02LAKK171 pKa = 9.78KK172 pKa = 10.45RR173 pKa = 11.84IVTPDD178 pKa = 3.5DD179 pKa = 3.56LVGEE183 pKa = 4.27RR184 pKa = 11.84LIGSPTDD191 pKa = 3.46TPYY194 pKa = 11.43GQALLRR200 pKa = 11.84AYY202 pKa = 9.84GPHH205 pKa = 7.13ASTLNVQSLVRR216 pKa = 11.84SATSACWQAQAGGGIAVVDD235 pKa = 4.66RR236 pKa = 11.84AAVAGTTFRR245 pKa = 11.84QLVVRR250 pKa = 11.84PFQTATRR257 pKa = 11.84LPIAVLHH264 pKa = 5.47NRR266 pKa = 11.84YY267 pKa = 9.4RR268 pKa = 11.84PLSVVQQAFCDD279 pKa = 3.4AFAAVWKK286 pKa = 10.6RR287 pKa = 11.84EE288 pKa = 3.95MPRR291 pKa = 3.2
MM1 pKa = 7.52NLRR4 pKa = 11.84HH5 pKa = 5.81MEE7 pKa = 3.9VFRR10 pKa = 11.84AVMLTGGVVGAAEE23 pKa = 4.88LLHH26 pKa = 6.64ISQPAVSKK34 pKa = 10.73ILAQAQRR41 pKa = 11.84QLGFALFVRR50 pKa = 11.84IKK52 pKa = 10.81GRR54 pKa = 11.84LVPTAEE60 pKa = 4.15GQALHH65 pKa = 7.01AEE67 pKa = 4.56VEE69 pKa = 4.58TLWRR73 pKa = 11.84SVQRR77 pKa = 11.84VQATPRR83 pKa = 11.84SGTLTLAVSASLAPYY98 pKa = 9.74LVPRR102 pKa = 11.84TLATLARR109 pKa = 11.84RR110 pKa = 11.84FPGLQTRR117 pKa = 11.84MEE119 pKa = 4.17ILVTPIMVDD128 pKa = 3.0ALLDD132 pKa = 3.5RR133 pKa = 11.84TADD136 pKa = 3.05IGVAILPNDD145 pKa = 3.92HH146 pKa = 7.28PNLVQVSRR154 pKa = 11.84HH155 pKa = 5.79DD156 pKa = 3.57CGFACVLPEE165 pKa = 3.8GHH167 pKa = 7.08KK168 pKa = 10.02LAKK171 pKa = 9.78KK172 pKa = 10.45RR173 pKa = 11.84IVTPDD178 pKa = 3.5DD179 pKa = 3.56LVGEE183 pKa = 4.27RR184 pKa = 11.84LIGSPTDD191 pKa = 3.46TPYY194 pKa = 11.43GQALLRR200 pKa = 11.84AYY202 pKa = 9.84GPHH205 pKa = 7.13ASTLNVQSLVRR216 pKa = 11.84SATSACWQAQAGGGIAVVDD235 pKa = 4.66RR236 pKa = 11.84AAVAGTTFRR245 pKa = 11.84QLVVRR250 pKa = 11.84PFQTATRR257 pKa = 11.84LPIAVLHH264 pKa = 5.47NRR266 pKa = 11.84YY267 pKa = 9.4RR268 pKa = 11.84PLSVVQQAFCDD279 pKa = 3.4AFAAVWKK286 pKa = 10.6RR287 pKa = 11.84EE288 pKa = 3.95MPRR291 pKa = 3.2
Molecular weight: 31.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1496469 |
24 |
1916 |
254.8 |
27.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.268 ± 0.036 | 0.949 ± 0.01 |
5.062 ± 0.026 | 4.856 ± 0.027 |
3.572 ± 0.02 | 8.649 ± 0.031 |
2.252 ± 0.016 | 4.234 ± 0.025 |
3.106 ± 0.027 | 10.817 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.552 ± 0.018 | 2.563 ± 0.02 |
5.366 ± 0.028 | 4.045 ± 0.021 |
6.637 ± 0.034 | 5.44 ± 0.025 |
5.278 ± 0.026 | 7.693 ± 0.029 |
1.481 ± 0.014 | 2.179 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
