
Mossman virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Narmovirus; Mossman narmovirus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
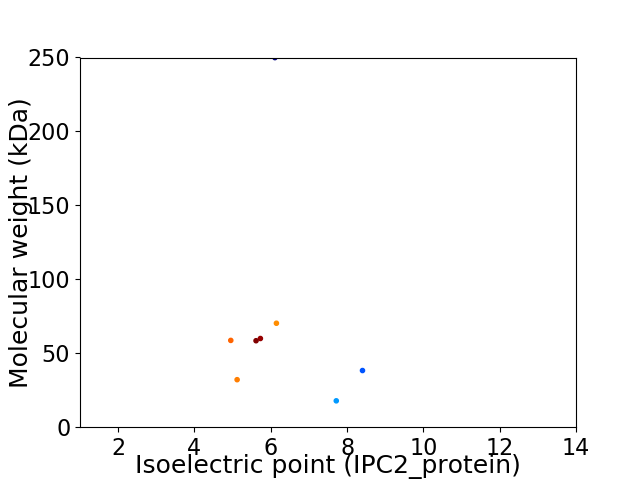
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6WGM3|Q6WGM3_9MONO Isoform of Q6WGM5 C protein OS=Mossman virus OX=241630 GN=C PE=4 SV=1
MM1 pKa = 7.39SGVLSALRR9 pKa = 11.84EE10 pKa = 4.13FKK12 pKa = 10.49DD13 pKa = 3.52ARR15 pKa = 11.84LASKK19 pKa = 11.28GEE21 pKa = 3.85GLTRR25 pKa = 11.84GAITGIKK32 pKa = 9.61QKK34 pKa = 10.2IAVIIPGQEE43 pKa = 3.68GSKK46 pKa = 10.52VRR48 pKa = 11.84WQLLRR53 pKa = 11.84LLLGVIWSQEE63 pKa = 3.61ASPGVITGAFLSLISLISEE82 pKa = 4.3SPGNMVRR89 pKa = 11.84RR90 pKa = 11.84LNNDD94 pKa = 3.08PDD96 pKa = 3.75LAITIIEE103 pKa = 4.19FTIGPDD109 pKa = 3.31SEE111 pKa = 4.9YY112 pKa = 11.13KK113 pKa = 9.53FASRR117 pKa = 11.84GMSYY121 pKa = 10.46EE122 pKa = 3.87EE123 pKa = 4.1QMAHH127 pKa = 5.78YY128 pKa = 9.87LHH130 pKa = 7.01LRR132 pKa = 11.84DD133 pKa = 4.76TPPQSAPDD141 pKa = 3.8DD142 pKa = 4.17FPFEE146 pKa = 4.07EE147 pKa = 4.46SEE149 pKa = 3.91AWRR152 pKa = 11.84QDD154 pKa = 3.73DD155 pKa = 3.67MPMDD159 pKa = 4.18EE160 pKa = 5.01FLVANMTVQVQLWTLLIKK178 pKa = 10.63AVTAPDD184 pKa = 3.53TARR187 pKa = 11.84DD188 pKa = 3.86GEE190 pKa = 4.08QRR192 pKa = 11.84RR193 pKa = 11.84WLKK196 pKa = 10.05FVQQRR201 pKa = 11.84RR202 pKa = 11.84VEE204 pKa = 4.27SFYY207 pKa = 11.4KK208 pKa = 10.63LHH210 pKa = 5.67TVWMDD215 pKa = 3.86RR216 pKa = 11.84ARR218 pKa = 11.84MHH220 pKa = 6.64IAASLSIRR228 pKa = 11.84RR229 pKa = 11.84YY230 pKa = 7.29MVKK233 pKa = 9.14TLIEE237 pKa = 4.17IQRR240 pKa = 11.84MGQGKK245 pKa = 9.37GRR247 pKa = 11.84LLEE250 pKa = 4.35VIADD254 pKa = 3.39IGNYY258 pKa = 9.27IEE260 pKa = 4.31EE261 pKa = 4.44SGLAGFQLTIRR272 pKa = 11.84FGIEE276 pKa = 3.36TKK278 pKa = 10.7YY279 pKa = 10.9AALALNEE286 pKa = 4.07FQGDD290 pKa = 3.14IATIEE295 pKa = 4.18RR296 pKa = 11.84LMKK299 pKa = 10.46LYY301 pKa = 11.08LEE303 pKa = 5.38LGPTAPFMVLLEE315 pKa = 4.94DD316 pKa = 4.9SIQTRR321 pKa = 11.84FAPGNYY327 pKa = 8.54PLLWSYY333 pKa = 12.11AMGVGSALDD342 pKa = 3.0RR343 pKa = 11.84AMANLNFNRR352 pKa = 11.84SYY354 pKa = 11.45LDD356 pKa = 2.93YY357 pKa = 11.02GYY359 pKa = 10.82FRR361 pKa = 11.84LGYY364 pKa = 9.65RR365 pKa = 11.84IVRR368 pKa = 11.84QSEE371 pKa = 4.36GSVDD375 pKa = 2.9TRR377 pKa = 11.84MARR380 pKa = 11.84EE381 pKa = 3.8LGITDD386 pKa = 4.02EE387 pKa = 4.24EE388 pKa = 4.34QQRR391 pKa = 11.84LRR393 pKa = 11.84RR394 pKa = 11.84LVADD398 pKa = 4.47LGNRR402 pKa = 11.84GDD404 pKa = 4.22SEE406 pKa = 3.95AAAYY410 pKa = 9.91QGGAFQLANIQDD422 pKa = 4.06FEE424 pKa = 5.02NDD426 pKa = 3.27DD427 pKa = 3.75AFAANPQQAPRR438 pKa = 11.84NNRR441 pKa = 11.84NRR443 pKa = 11.84RR444 pKa = 11.84RR445 pKa = 11.84RR446 pKa = 11.84GQPDD450 pKa = 2.97QGDD453 pKa = 4.19DD454 pKa = 3.75DD455 pKa = 6.25DD456 pKa = 6.45SDD458 pKa = 3.99EE459 pKa = 5.73GDD461 pKa = 3.66DD462 pKa = 4.43QGGSGYY468 pKa = 10.67AAAVHH473 pKa = 6.24AVLHH477 pKa = 5.95SSGDD481 pKa = 3.55GGDD484 pKa = 4.49DD485 pKa = 3.54LTSGADD491 pKa = 3.69GSIDD495 pKa = 3.76PPTSGLGANYY505 pKa = 9.92RR506 pKa = 11.84YY507 pKa = 10.45SPGLNDD513 pKa = 4.82SKK515 pKa = 11.32PKK517 pKa = 10.78FNDD520 pKa = 3.04ADD522 pKa = 4.19LLGLGDD528 pKa = 3.76
MM1 pKa = 7.39SGVLSALRR9 pKa = 11.84EE10 pKa = 4.13FKK12 pKa = 10.49DD13 pKa = 3.52ARR15 pKa = 11.84LASKK19 pKa = 11.28GEE21 pKa = 3.85GLTRR25 pKa = 11.84GAITGIKK32 pKa = 9.61QKK34 pKa = 10.2IAVIIPGQEE43 pKa = 3.68GSKK46 pKa = 10.52VRR48 pKa = 11.84WQLLRR53 pKa = 11.84LLLGVIWSQEE63 pKa = 3.61ASPGVITGAFLSLISLISEE82 pKa = 4.3SPGNMVRR89 pKa = 11.84RR90 pKa = 11.84LNNDD94 pKa = 3.08PDD96 pKa = 3.75LAITIIEE103 pKa = 4.19FTIGPDD109 pKa = 3.31SEE111 pKa = 4.9YY112 pKa = 11.13KK113 pKa = 9.53FASRR117 pKa = 11.84GMSYY121 pKa = 10.46EE122 pKa = 3.87EE123 pKa = 4.1QMAHH127 pKa = 5.78YY128 pKa = 9.87LHH130 pKa = 7.01LRR132 pKa = 11.84DD133 pKa = 4.76TPPQSAPDD141 pKa = 3.8DD142 pKa = 4.17FPFEE146 pKa = 4.07EE147 pKa = 4.46SEE149 pKa = 3.91AWRR152 pKa = 11.84QDD154 pKa = 3.73DD155 pKa = 3.67MPMDD159 pKa = 4.18EE160 pKa = 5.01FLVANMTVQVQLWTLLIKK178 pKa = 10.63AVTAPDD184 pKa = 3.53TARR187 pKa = 11.84DD188 pKa = 3.86GEE190 pKa = 4.08QRR192 pKa = 11.84RR193 pKa = 11.84WLKK196 pKa = 10.05FVQQRR201 pKa = 11.84RR202 pKa = 11.84VEE204 pKa = 4.27SFYY207 pKa = 11.4KK208 pKa = 10.63LHH210 pKa = 5.67TVWMDD215 pKa = 3.86RR216 pKa = 11.84ARR218 pKa = 11.84MHH220 pKa = 6.64IAASLSIRR228 pKa = 11.84RR229 pKa = 11.84YY230 pKa = 7.29MVKK233 pKa = 9.14TLIEE237 pKa = 4.17IQRR240 pKa = 11.84MGQGKK245 pKa = 9.37GRR247 pKa = 11.84LLEE250 pKa = 4.35VIADD254 pKa = 3.39IGNYY258 pKa = 9.27IEE260 pKa = 4.31EE261 pKa = 4.44SGLAGFQLTIRR272 pKa = 11.84FGIEE276 pKa = 3.36TKK278 pKa = 10.7YY279 pKa = 10.9AALALNEE286 pKa = 4.07FQGDD290 pKa = 3.14IATIEE295 pKa = 4.18RR296 pKa = 11.84LMKK299 pKa = 10.46LYY301 pKa = 11.08LEE303 pKa = 5.38LGPTAPFMVLLEE315 pKa = 4.94DD316 pKa = 4.9SIQTRR321 pKa = 11.84FAPGNYY327 pKa = 8.54PLLWSYY333 pKa = 12.11AMGVGSALDD342 pKa = 3.0RR343 pKa = 11.84AMANLNFNRR352 pKa = 11.84SYY354 pKa = 11.45LDD356 pKa = 2.93YY357 pKa = 11.02GYY359 pKa = 10.82FRR361 pKa = 11.84LGYY364 pKa = 9.65RR365 pKa = 11.84IVRR368 pKa = 11.84QSEE371 pKa = 4.36GSVDD375 pKa = 2.9TRR377 pKa = 11.84MARR380 pKa = 11.84EE381 pKa = 3.8LGITDD386 pKa = 4.02EE387 pKa = 4.24EE388 pKa = 4.34QQRR391 pKa = 11.84LRR393 pKa = 11.84RR394 pKa = 11.84LVADD398 pKa = 4.47LGNRR402 pKa = 11.84GDD404 pKa = 4.22SEE406 pKa = 3.95AAAYY410 pKa = 9.91QGGAFQLANIQDD422 pKa = 4.06FEE424 pKa = 5.02NDD426 pKa = 3.27DD427 pKa = 3.75AFAANPQQAPRR438 pKa = 11.84NNRR441 pKa = 11.84NRR443 pKa = 11.84RR444 pKa = 11.84RR445 pKa = 11.84RR446 pKa = 11.84GQPDD450 pKa = 2.97QGDD453 pKa = 4.19DD454 pKa = 3.75DD455 pKa = 6.25DD456 pKa = 6.45SDD458 pKa = 3.99EE459 pKa = 5.73GDD461 pKa = 3.66DD462 pKa = 4.43QGGSGYY468 pKa = 10.67AAAVHH473 pKa = 6.24AVLHH477 pKa = 5.95SSGDD481 pKa = 3.55GGDD484 pKa = 4.49DD485 pKa = 3.54LTSGADD491 pKa = 3.69GSIDD495 pKa = 3.76PPTSGLGANYY505 pKa = 9.92RR506 pKa = 11.84YY507 pKa = 10.45SPGLNDD513 pKa = 4.82SKK515 pKa = 11.32PKK517 pKa = 10.78FNDD520 pKa = 3.04ADD522 pKa = 4.19LLGLGDD528 pKa = 3.76
Molecular weight: 58.64 kDa
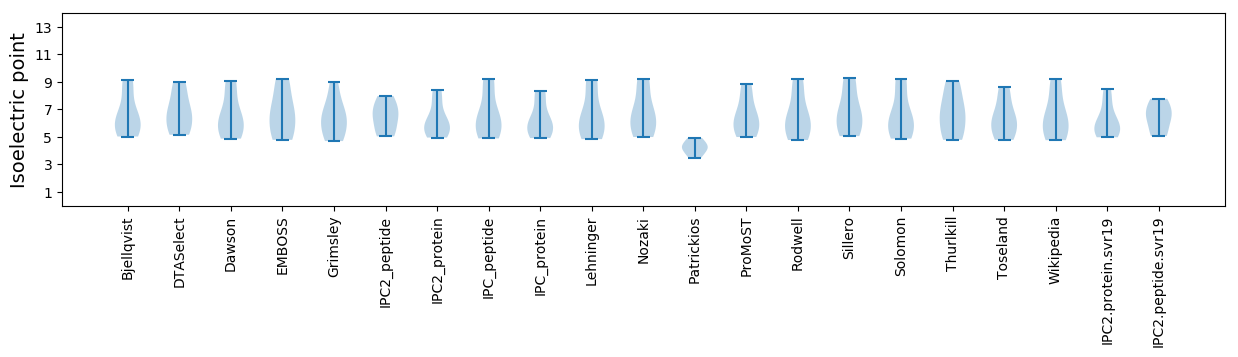
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6WGM5|Q6WGM5_9MONO Phosphoprotein OS=Mossman virus OX=241630 GN=C PE=4 SV=1
MM1 pKa = 7.15ATMHH5 pKa = 6.49EE6 pKa = 4.7FLPGTWVNRR15 pKa = 11.84GLLEE19 pKa = 5.16DD20 pKa = 3.88IRR22 pKa = 11.84PEE24 pKa = 3.78YY25 pKa = 10.6DD26 pKa = 2.95HH27 pKa = 7.03EE28 pKa = 4.74GKK30 pKa = 10.13IKK32 pKa = 10.35PRR34 pKa = 11.84VRR36 pKa = 11.84VIDD39 pKa = 4.02PGAGTRR45 pKa = 11.84KK46 pKa = 10.05SSGFMYY52 pKa = 10.52LHH54 pKa = 6.31LQGIIEE60 pKa = 4.15DD61 pKa = 4.16WIGDD65 pKa = 3.85FEE67 pKa = 4.74PATRR71 pKa = 11.84PPGRR75 pKa = 11.84TLAAYY80 pKa = 8.55PLGVGQSIAGPYY92 pKa = 9.53EE93 pKa = 4.28LLTACLEE100 pKa = 4.29LNVVVRR106 pKa = 11.84RR107 pKa = 11.84TVGSTEE113 pKa = 3.66KK114 pKa = 10.91VVFYY118 pKa = 11.35NNAALDD124 pKa = 3.81VLSPWKK130 pKa = 10.13NVLMNGCIFDD140 pKa = 3.87ANKK143 pKa = 8.87VCKK146 pKa = 9.78RR147 pKa = 11.84VEE149 pKa = 4.69DD150 pKa = 3.57IPLEE154 pKa = 4.1KK155 pKa = 10.11EE156 pKa = 3.69QRR158 pKa = 11.84FRR160 pKa = 11.84PIYY163 pKa = 9.55LTITLLTDD171 pKa = 3.08SGLYY175 pKa = 7.73KK176 pKa = 9.81TPSMIQDD183 pKa = 2.95IRR185 pKa = 11.84ANNAVAFNLLVYY197 pKa = 8.57LTAGNGIMEE206 pKa = 4.88KK207 pKa = 10.18LYY209 pKa = 9.81PHH211 pKa = 7.02TGSEE215 pKa = 4.32PEE217 pKa = 3.9QTVVSFMVHH226 pKa = 5.06VGLFMRR232 pKa = 11.84GRR234 pKa = 11.84KK235 pKa = 8.78KK236 pKa = 10.57AYY238 pKa = 9.08SHH240 pKa = 7.34DD241 pKa = 3.74YY242 pKa = 10.74CRR244 pKa = 11.84LKK246 pKa = 10.62VEE248 pKa = 4.45RR249 pKa = 11.84MNLQFGLGGVGGVSFHH265 pKa = 6.32MRR267 pKa = 11.84VQGKK271 pKa = 9.45LSKK274 pKa = 10.44SLHH277 pKa = 5.67AQLGFHH283 pKa = 7.16RR284 pKa = 11.84SVCYY288 pKa = 10.67SLMDD292 pKa = 4.56INPALNKK299 pKa = 10.23LLWRR303 pKa = 11.84TQCSIQRR310 pKa = 11.84VVAVFQPSVPSEE322 pKa = 3.33FRR324 pKa = 11.84IYY326 pKa = 11.23NDD328 pKa = 3.15VLIDD332 pKa = 3.62NTGKK336 pKa = 10.49IMTYY340 pKa = 10.69
MM1 pKa = 7.15ATMHH5 pKa = 6.49EE6 pKa = 4.7FLPGTWVNRR15 pKa = 11.84GLLEE19 pKa = 5.16DD20 pKa = 3.88IRR22 pKa = 11.84PEE24 pKa = 3.78YY25 pKa = 10.6DD26 pKa = 2.95HH27 pKa = 7.03EE28 pKa = 4.74GKK30 pKa = 10.13IKK32 pKa = 10.35PRR34 pKa = 11.84VRR36 pKa = 11.84VIDD39 pKa = 4.02PGAGTRR45 pKa = 11.84KK46 pKa = 10.05SSGFMYY52 pKa = 10.52LHH54 pKa = 6.31LQGIIEE60 pKa = 4.15DD61 pKa = 4.16WIGDD65 pKa = 3.85FEE67 pKa = 4.74PATRR71 pKa = 11.84PPGRR75 pKa = 11.84TLAAYY80 pKa = 8.55PLGVGQSIAGPYY92 pKa = 9.53EE93 pKa = 4.28LLTACLEE100 pKa = 4.29LNVVVRR106 pKa = 11.84RR107 pKa = 11.84TVGSTEE113 pKa = 3.66KK114 pKa = 10.91VVFYY118 pKa = 11.35NNAALDD124 pKa = 3.81VLSPWKK130 pKa = 10.13NVLMNGCIFDD140 pKa = 3.87ANKK143 pKa = 8.87VCKK146 pKa = 9.78RR147 pKa = 11.84VEE149 pKa = 4.69DD150 pKa = 3.57IPLEE154 pKa = 4.1KK155 pKa = 10.11EE156 pKa = 3.69QRR158 pKa = 11.84FRR160 pKa = 11.84PIYY163 pKa = 9.55LTITLLTDD171 pKa = 3.08SGLYY175 pKa = 7.73KK176 pKa = 9.81TPSMIQDD183 pKa = 2.95IRR185 pKa = 11.84ANNAVAFNLLVYY197 pKa = 8.57LTAGNGIMEE206 pKa = 4.88KK207 pKa = 10.18LYY209 pKa = 9.81PHH211 pKa = 7.02TGSEE215 pKa = 4.32PEE217 pKa = 3.9QTVVSFMVHH226 pKa = 5.06VGLFMRR232 pKa = 11.84GRR234 pKa = 11.84KK235 pKa = 8.78KK236 pKa = 10.57AYY238 pKa = 9.08SHH240 pKa = 7.34DD241 pKa = 3.74YY242 pKa = 10.74CRR244 pKa = 11.84LKK246 pKa = 10.62VEE248 pKa = 4.45RR249 pKa = 11.84MNLQFGLGGVGGVSFHH265 pKa = 6.32MRR267 pKa = 11.84VQGKK271 pKa = 9.45LSKK274 pKa = 10.44SLHH277 pKa = 5.67AQLGFHH283 pKa = 7.16RR284 pKa = 11.84SVCYY288 pKa = 10.67SLMDD292 pKa = 4.56INPALNKK299 pKa = 10.23LLWRR303 pKa = 11.84TQCSIQRR310 pKa = 11.84VVAVFQPSVPSEE322 pKa = 3.33FRR324 pKa = 11.84IYY326 pKa = 11.23NDD328 pKa = 3.15VLIDD332 pKa = 3.62NTGKK336 pKa = 10.49IMTYY340 pKa = 10.69
Molecular weight: 38.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5246 |
152 |
2205 |
655.8 |
73.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.538 ± 0.43 | 1.639 ± 0.303 |
5.471 ± 0.349 | 5.623 ± 0.601 |
2.993 ± 0.409 | 6.348 ± 0.721 |
2.402 ± 0.506 | 7.301 ± 0.743 |
4.727 ± 0.366 | 9.379 ± 0.622 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.592 ± 0.213 | 5.032 ± 0.311 |
4.918 ± 0.211 | 4.175 ± 0.412 |
5.49 ± 0.465 | 8.254 ± 0.678 |
6.271 ± 0.641 | 6.176 ± 0.497 |
1.106 ± 0.186 | 3.565 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
