
Asparagus virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 7.42
Get precalculated fractions of proteins
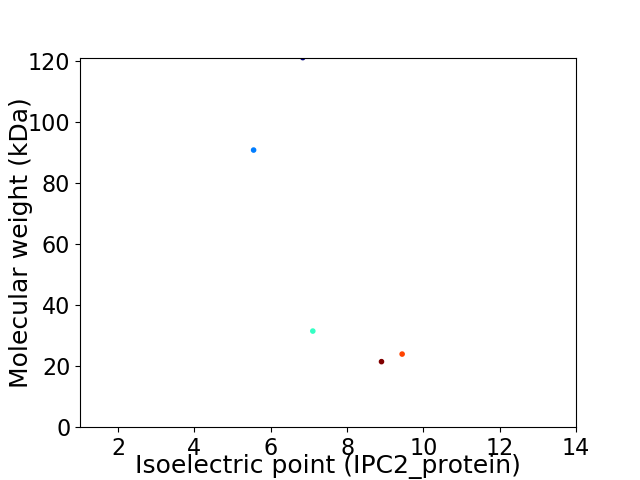
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B8RCA5|B8RCA5_9BROM 2b protein OS=Asparagus virus 2 OX=39681 GN=2b PE=4 SV=1
MM1 pKa = 7.91DD2 pKa = 3.62AHH4 pKa = 6.46YY5 pKa = 9.92FLSEE9 pKa = 3.86LCDD12 pKa = 3.05IRR14 pKa = 11.84KK15 pKa = 8.66FVSTRR20 pKa = 11.84VIEE23 pKa = 3.91QSFDD27 pKa = 3.26IKK29 pKa = 11.08DD30 pKa = 4.1DD31 pKa = 3.54EE32 pKa = 4.68TTSYY36 pKa = 10.05LQAWMYY42 pKa = 11.19SRR44 pKa = 11.84ILVPVMKK51 pKa = 10.27FHH53 pKa = 7.66LCGEE57 pKa = 4.21VLLDD61 pKa = 4.33KK62 pKa = 11.03IEE64 pKa = 4.36DD65 pKa = 3.79VFRR68 pKa = 11.84EE69 pKa = 4.11TLGCEE74 pKa = 4.33AEE76 pKa = 4.4PNDD79 pKa = 4.6DD80 pKa = 3.22GYY82 pKa = 11.1MPVFIHH88 pKa = 7.44LDD90 pKa = 3.74DD91 pKa = 6.5DD92 pKa = 5.06DD93 pKa = 6.12DD94 pKa = 4.39EE95 pKa = 5.29PPPEE99 pKa = 4.93AYY101 pKa = 10.14LPKK104 pKa = 10.23EE105 pKa = 3.87EE106 pKa = 5.0RR107 pKa = 11.84EE108 pKa = 4.47AIHH111 pKa = 6.68AATFLGHH118 pKa = 5.63EE119 pKa = 4.36HH120 pKa = 6.52WASVTSGSDD129 pKa = 3.25FYY131 pKa = 11.91DD132 pKa = 4.94DD133 pKa = 4.71IEE135 pKa = 4.25MLADD139 pKa = 3.27EE140 pKa = 5.39TIYY143 pKa = 11.49DD144 pKa = 4.16DD145 pKa = 5.31LQTDD149 pKa = 3.75KK150 pKa = 10.81CAEE153 pKa = 4.07MPADD157 pKa = 4.06YY158 pKa = 10.89QIMWNDD164 pKa = 3.57GAIDD168 pKa = 4.21AVWSSQFEE176 pKa = 4.49CEE178 pKa = 4.45EE179 pKa = 3.97PPKK182 pKa = 10.65PKK184 pKa = 10.4FKK186 pKa = 10.19PQSVDD191 pKa = 2.99KK192 pKa = 11.15GCDD195 pKa = 3.21PVVIQAAIDD204 pKa = 4.38DD205 pKa = 4.42IFPFHH210 pKa = 7.96HH211 pKa = 6.58EE212 pKa = 3.42MDD214 pKa = 3.33DD215 pKa = 4.56RR216 pKa = 11.84YY217 pKa = 10.34FQTLVEE223 pKa = 4.32TQDD226 pKa = 2.99ISLEE230 pKa = 4.17VSKK233 pKa = 10.97CWIDD237 pKa = 3.22ASNFRR242 pKa = 11.84DD243 pKa = 4.1FVKK246 pKa = 10.9GQDD249 pKa = 3.44SYY251 pKa = 11.95AVPVIQSGATSKK263 pKa = 10.43RR264 pKa = 11.84VHH266 pKa = 5.39TQRR269 pKa = 11.84EE270 pKa = 3.7ALLAVKK276 pKa = 9.4KK277 pKa = 10.59RR278 pKa = 11.84NMNIPEE284 pKa = 4.22LQSTFDD290 pKa = 4.64LDD292 pKa = 4.98AEE294 pKa = 4.56VNTCFKK300 pKa = 10.81RR301 pKa = 11.84FLTHH305 pKa = 6.18VVDD308 pKa = 4.24VPRR311 pKa = 11.84LRR313 pKa = 11.84RR314 pKa = 11.84LQPMTGVEE322 pKa = 4.04VEE324 pKa = 4.59FFNQYY329 pKa = 11.22LLGKK333 pKa = 9.63NPPLKK338 pKa = 10.01EE339 pKa = 4.06YY340 pKa = 10.53QGPLPLCSLDD350 pKa = 4.25KK351 pKa = 10.73YY352 pKa = 9.09MHH354 pKa = 5.51MVKK357 pKa = 9.98TIVKK361 pKa = 8.75PVEE364 pKa = 4.43DD365 pKa = 3.85NSLKK369 pKa = 10.61YY370 pKa = 10.06EE371 pKa = 4.23RR372 pKa = 11.84PLCATITYY380 pKa = 8.26HH381 pKa = 6.97KK382 pKa = 9.85KK383 pKa = 10.32GIVMQSSPLFLSAMSRR399 pKa = 11.84LFYY402 pKa = 10.61VLKK405 pKa = 10.64SKK407 pKa = 10.19IHH409 pKa = 5.86IPSGKK414 pKa = 7.2WHH416 pKa = 6.3QLFTLDD422 pKa = 5.07AEE424 pKa = 4.7KK425 pKa = 10.61FDD427 pKa = 3.55AAKK430 pKa = 9.69WFKK433 pKa = 10.72EE434 pKa = 3.7VDD436 pKa = 3.71FSKK439 pKa = 10.64FDD441 pKa = 3.3KK442 pKa = 11.19SQGEE446 pKa = 4.14LHH448 pKa = 6.89HH449 pKa = 6.75KK450 pKa = 8.67VQEE453 pKa = 4.67KK454 pKa = 10.38IFEE457 pKa = 4.32CLKK460 pKa = 10.44LPPGFVEE467 pKa = 4.1MWFTALEE474 pKa = 4.04RR475 pKa = 11.84SHH477 pKa = 8.3IVDD480 pKa = 3.93RR481 pKa = 11.84EE482 pKa = 3.95AGIGFSVDD490 pKa = 3.65FQRR493 pKa = 11.84RR494 pKa = 11.84TGDD497 pKa = 3.07ANTYY501 pKa = 10.16LGNTLVTLICLARR514 pKa = 11.84VYY516 pKa = 10.41DD517 pKa = 4.03LCNPNITFVIASGDD531 pKa = 3.51DD532 pKa = 3.61SLIGSVDD539 pKa = 3.48EE540 pKa = 4.67LPRR543 pKa = 11.84GPEE546 pKa = 3.88DD547 pKa = 4.79LFTTLFNFEE556 pKa = 4.52AKK558 pKa = 10.1FPHH561 pKa = 5.93NQPFICSKK569 pKa = 10.53FLVSVDD575 pKa = 3.54LADD578 pKa = 4.2GGRR581 pKa = 11.84EE582 pKa = 4.11VIAVPNPAKK591 pKa = 10.65LLIRR595 pKa = 11.84MGRR598 pKa = 11.84RR599 pKa = 11.84DD600 pKa = 3.97CQYY603 pKa = 10.86QALDD607 pKa = 3.78DD608 pKa = 5.92LYY610 pKa = 10.84TSWLDD615 pKa = 3.25VIYY618 pKa = 10.19YY619 pKa = 10.13FRR621 pKa = 11.84DD622 pKa = 3.06SRR624 pKa = 11.84VCEE627 pKa = 4.17KK628 pKa = 10.72VASLCAFRR636 pKa = 11.84QTRR639 pKa = 11.84RR640 pKa = 11.84SSMYY644 pKa = 10.48LLSALLSLPSRR655 pKa = 11.84FANKK659 pKa = 10.05KK660 pKa = 8.24KK661 pKa = 10.66FKK663 pKa = 8.91WLCYY667 pKa = 9.73HH668 pKa = 6.61LTEE671 pKa = 4.61QEE673 pKa = 4.14CLKK676 pKa = 9.5ITKK679 pKa = 9.95QPGSSHH685 pKa = 4.08YY686 pKa = 9.02TKK688 pKa = 10.76KK689 pKa = 10.72NEE691 pKa = 4.02CSKK694 pKa = 10.9EE695 pKa = 4.19SVKK698 pKa = 10.76NFPHH702 pKa = 6.62RR703 pKa = 11.84RR704 pKa = 11.84SSCGRR709 pKa = 11.84WEE711 pKa = 5.18FIPKK715 pKa = 9.96IKK717 pKa = 10.64SDD719 pKa = 3.46DD720 pKa = 3.84DD721 pKa = 3.52YY722 pKa = 11.72RR723 pKa = 11.84IFPNGDD729 pKa = 3.02LSTIRR734 pKa = 11.84GSSVSKK740 pKa = 9.72TDD742 pKa = 3.31CGYY745 pKa = 11.31GKK747 pKa = 10.76CSDD750 pKa = 3.74TASSIGNKK758 pKa = 6.8NTRR761 pKa = 11.84PRR763 pKa = 11.84NRR765 pKa = 11.84FRR767 pKa = 11.84AEE769 pKa = 3.98GVHH772 pKa = 5.76QPSGCYY778 pKa = 9.66SNRR781 pKa = 11.84LPPNHH786 pKa = 6.59RR787 pKa = 11.84RR788 pKa = 11.84SSS790 pKa = 3.3
MM1 pKa = 7.91DD2 pKa = 3.62AHH4 pKa = 6.46YY5 pKa = 9.92FLSEE9 pKa = 3.86LCDD12 pKa = 3.05IRR14 pKa = 11.84KK15 pKa = 8.66FVSTRR20 pKa = 11.84VIEE23 pKa = 3.91QSFDD27 pKa = 3.26IKK29 pKa = 11.08DD30 pKa = 4.1DD31 pKa = 3.54EE32 pKa = 4.68TTSYY36 pKa = 10.05LQAWMYY42 pKa = 11.19SRR44 pKa = 11.84ILVPVMKK51 pKa = 10.27FHH53 pKa = 7.66LCGEE57 pKa = 4.21VLLDD61 pKa = 4.33KK62 pKa = 11.03IEE64 pKa = 4.36DD65 pKa = 3.79VFRR68 pKa = 11.84EE69 pKa = 4.11TLGCEE74 pKa = 4.33AEE76 pKa = 4.4PNDD79 pKa = 4.6DD80 pKa = 3.22GYY82 pKa = 11.1MPVFIHH88 pKa = 7.44LDD90 pKa = 3.74DD91 pKa = 6.5DD92 pKa = 5.06DD93 pKa = 6.12DD94 pKa = 4.39EE95 pKa = 5.29PPPEE99 pKa = 4.93AYY101 pKa = 10.14LPKK104 pKa = 10.23EE105 pKa = 3.87EE106 pKa = 5.0RR107 pKa = 11.84EE108 pKa = 4.47AIHH111 pKa = 6.68AATFLGHH118 pKa = 5.63EE119 pKa = 4.36HH120 pKa = 6.52WASVTSGSDD129 pKa = 3.25FYY131 pKa = 11.91DD132 pKa = 4.94DD133 pKa = 4.71IEE135 pKa = 4.25MLADD139 pKa = 3.27EE140 pKa = 5.39TIYY143 pKa = 11.49DD144 pKa = 4.16DD145 pKa = 5.31LQTDD149 pKa = 3.75KK150 pKa = 10.81CAEE153 pKa = 4.07MPADD157 pKa = 4.06YY158 pKa = 10.89QIMWNDD164 pKa = 3.57GAIDD168 pKa = 4.21AVWSSQFEE176 pKa = 4.49CEE178 pKa = 4.45EE179 pKa = 3.97PPKK182 pKa = 10.65PKK184 pKa = 10.4FKK186 pKa = 10.19PQSVDD191 pKa = 2.99KK192 pKa = 11.15GCDD195 pKa = 3.21PVVIQAAIDD204 pKa = 4.38DD205 pKa = 4.42IFPFHH210 pKa = 7.96HH211 pKa = 6.58EE212 pKa = 3.42MDD214 pKa = 3.33DD215 pKa = 4.56RR216 pKa = 11.84YY217 pKa = 10.34FQTLVEE223 pKa = 4.32TQDD226 pKa = 2.99ISLEE230 pKa = 4.17VSKK233 pKa = 10.97CWIDD237 pKa = 3.22ASNFRR242 pKa = 11.84DD243 pKa = 4.1FVKK246 pKa = 10.9GQDD249 pKa = 3.44SYY251 pKa = 11.95AVPVIQSGATSKK263 pKa = 10.43RR264 pKa = 11.84VHH266 pKa = 5.39TQRR269 pKa = 11.84EE270 pKa = 3.7ALLAVKK276 pKa = 9.4KK277 pKa = 10.59RR278 pKa = 11.84NMNIPEE284 pKa = 4.22LQSTFDD290 pKa = 4.64LDD292 pKa = 4.98AEE294 pKa = 4.56VNTCFKK300 pKa = 10.81RR301 pKa = 11.84FLTHH305 pKa = 6.18VVDD308 pKa = 4.24VPRR311 pKa = 11.84LRR313 pKa = 11.84RR314 pKa = 11.84LQPMTGVEE322 pKa = 4.04VEE324 pKa = 4.59FFNQYY329 pKa = 11.22LLGKK333 pKa = 9.63NPPLKK338 pKa = 10.01EE339 pKa = 4.06YY340 pKa = 10.53QGPLPLCSLDD350 pKa = 4.25KK351 pKa = 10.73YY352 pKa = 9.09MHH354 pKa = 5.51MVKK357 pKa = 9.98TIVKK361 pKa = 8.75PVEE364 pKa = 4.43DD365 pKa = 3.85NSLKK369 pKa = 10.61YY370 pKa = 10.06EE371 pKa = 4.23RR372 pKa = 11.84PLCATITYY380 pKa = 8.26HH381 pKa = 6.97KK382 pKa = 9.85KK383 pKa = 10.32GIVMQSSPLFLSAMSRR399 pKa = 11.84LFYY402 pKa = 10.61VLKK405 pKa = 10.64SKK407 pKa = 10.19IHH409 pKa = 5.86IPSGKK414 pKa = 7.2WHH416 pKa = 6.3QLFTLDD422 pKa = 5.07AEE424 pKa = 4.7KK425 pKa = 10.61FDD427 pKa = 3.55AAKK430 pKa = 9.69WFKK433 pKa = 10.72EE434 pKa = 3.7VDD436 pKa = 3.71FSKK439 pKa = 10.64FDD441 pKa = 3.3KK442 pKa = 11.19SQGEE446 pKa = 4.14LHH448 pKa = 6.89HH449 pKa = 6.75KK450 pKa = 8.67VQEE453 pKa = 4.67KK454 pKa = 10.38IFEE457 pKa = 4.32CLKK460 pKa = 10.44LPPGFVEE467 pKa = 4.1MWFTALEE474 pKa = 4.04RR475 pKa = 11.84SHH477 pKa = 8.3IVDD480 pKa = 3.93RR481 pKa = 11.84EE482 pKa = 3.95AGIGFSVDD490 pKa = 3.65FQRR493 pKa = 11.84RR494 pKa = 11.84TGDD497 pKa = 3.07ANTYY501 pKa = 10.16LGNTLVTLICLARR514 pKa = 11.84VYY516 pKa = 10.41DD517 pKa = 4.03LCNPNITFVIASGDD531 pKa = 3.51DD532 pKa = 3.61SLIGSVDD539 pKa = 3.48EE540 pKa = 4.67LPRR543 pKa = 11.84GPEE546 pKa = 3.88DD547 pKa = 4.79LFTTLFNFEE556 pKa = 4.52AKK558 pKa = 10.1FPHH561 pKa = 5.93NQPFICSKK569 pKa = 10.53FLVSVDD575 pKa = 3.54LADD578 pKa = 4.2GGRR581 pKa = 11.84EE582 pKa = 4.11VIAVPNPAKK591 pKa = 10.65LLIRR595 pKa = 11.84MGRR598 pKa = 11.84RR599 pKa = 11.84DD600 pKa = 3.97CQYY603 pKa = 10.86QALDD607 pKa = 3.78DD608 pKa = 5.92LYY610 pKa = 10.84TSWLDD615 pKa = 3.25VIYY618 pKa = 10.19YY619 pKa = 10.13FRR621 pKa = 11.84DD622 pKa = 3.06SRR624 pKa = 11.84VCEE627 pKa = 4.17KK628 pKa = 10.72VASLCAFRR636 pKa = 11.84QTRR639 pKa = 11.84RR640 pKa = 11.84SSMYY644 pKa = 10.48LLSALLSLPSRR655 pKa = 11.84FANKK659 pKa = 10.05KK660 pKa = 8.24KK661 pKa = 10.66FKK663 pKa = 8.91WLCYY667 pKa = 9.73HH668 pKa = 6.61LTEE671 pKa = 4.61QEE673 pKa = 4.14CLKK676 pKa = 9.5ITKK679 pKa = 9.95QPGSSHH685 pKa = 4.08YY686 pKa = 9.02TKK688 pKa = 10.76KK689 pKa = 10.72NEE691 pKa = 4.02CSKK694 pKa = 10.9EE695 pKa = 4.19SVKK698 pKa = 10.76NFPHH702 pKa = 6.62RR703 pKa = 11.84RR704 pKa = 11.84SSCGRR709 pKa = 11.84WEE711 pKa = 5.18FIPKK715 pKa = 9.96IKK717 pKa = 10.64SDD719 pKa = 3.46DD720 pKa = 3.84DD721 pKa = 3.52YY722 pKa = 11.72RR723 pKa = 11.84IFPNGDD729 pKa = 3.02LSTIRR734 pKa = 11.84GSSVSKK740 pKa = 9.72TDD742 pKa = 3.31CGYY745 pKa = 11.31GKK747 pKa = 10.76CSDD750 pKa = 3.74TASSIGNKK758 pKa = 6.8NTRR761 pKa = 11.84PRR763 pKa = 11.84NRR765 pKa = 11.84FRR767 pKa = 11.84AEE769 pKa = 3.98GVHH772 pKa = 5.76QPSGCYY778 pKa = 9.66SNRR781 pKa = 11.84LPPNHH786 pKa = 6.59RR787 pKa = 11.84RR788 pKa = 11.84SSS790 pKa = 3.3
Molecular weight: 90.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q65299|Q65299_9BROM Coat protein OS=Asparagus virus 2 OX=39681 GN=ORF2 PE=3 SV=1
MM1 pKa = 7.53SGNAIEE7 pKa = 5.15VNGRR11 pKa = 11.84WYY13 pKa = 9.75TPAPNNAPTRR23 pKa = 11.84GRR25 pKa = 11.84GGRR28 pKa = 11.84RR29 pKa = 11.84QPTARR34 pKa = 11.84SRR36 pKa = 11.84QWAQGQANARR46 pKa = 11.84RR47 pKa = 11.84SQPQALMIGSVPTSLPTWKK66 pKa = 10.28SFPGEE71 pKa = 3.73QWHH74 pKa = 6.02EE75 pKa = 3.77VSGYY79 pKa = 10.36SFPNSWGSGTLAWLTMANEE98 pKa = 4.02LNKK101 pKa = 10.19IKK103 pKa = 9.63TLHH106 pKa = 6.71DD107 pKa = 3.35TTKK110 pKa = 10.29VYY112 pKa = 11.23SVMLGFTCNYY122 pKa = 9.62DD123 pKa = 3.24GYY125 pKa = 11.09AGFVEE130 pKa = 5.47GFSTSNQIHH139 pKa = 7.35PIAPDD144 pKa = 3.97RR145 pKa = 11.84IRR147 pKa = 11.84VKK149 pKa = 10.22KK150 pKa = 10.01GKK152 pKa = 10.24YY153 pKa = 8.57GARR156 pKa = 11.84QQVFPTGTTVSEE168 pKa = 4.53VRR170 pKa = 11.84SNWTFVWNFDD180 pKa = 3.53AAPPTGSKK188 pKa = 10.65DD189 pKa = 3.39AITVTKK195 pKa = 9.51FYY197 pKa = 10.44IATSPLPGVKK207 pKa = 9.77PPSNFLVVEE216 pKa = 4.35EE217 pKa = 4.43
MM1 pKa = 7.53SGNAIEE7 pKa = 5.15VNGRR11 pKa = 11.84WYY13 pKa = 9.75TPAPNNAPTRR23 pKa = 11.84GRR25 pKa = 11.84GGRR28 pKa = 11.84RR29 pKa = 11.84QPTARR34 pKa = 11.84SRR36 pKa = 11.84QWAQGQANARR46 pKa = 11.84RR47 pKa = 11.84SQPQALMIGSVPTSLPTWKK66 pKa = 10.28SFPGEE71 pKa = 3.73QWHH74 pKa = 6.02EE75 pKa = 3.77VSGYY79 pKa = 10.36SFPNSWGSGTLAWLTMANEE98 pKa = 4.02LNKK101 pKa = 10.19IKK103 pKa = 9.63TLHH106 pKa = 6.71DD107 pKa = 3.35TTKK110 pKa = 10.29VYY112 pKa = 11.23SVMLGFTCNYY122 pKa = 9.62DD123 pKa = 3.24GYY125 pKa = 11.09AGFVEE130 pKa = 5.47GFSTSNQIHH139 pKa = 7.35PIAPDD144 pKa = 3.97RR145 pKa = 11.84IRR147 pKa = 11.84VKK149 pKa = 10.22KK150 pKa = 10.01GKK152 pKa = 10.24YY153 pKa = 8.57GARR156 pKa = 11.84QQVFPTGTTVSEE168 pKa = 4.53VRR170 pKa = 11.84SNWTFVWNFDD180 pKa = 3.53AAPPTGSKK188 pKa = 10.65DD189 pKa = 3.39AITVTKK195 pKa = 9.51FYY197 pKa = 10.44IATSPLPGVKK207 pKa = 9.77PPSNFLVVEE216 pKa = 4.35EE217 pKa = 4.43
Molecular weight: 23.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2557 |
193 |
1076 |
511.4 |
57.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.118 ± 0.683 | 1.995 ± 0.496 |
6.844 ± 0.888 | 5.514 ± 0.504 |
4.654 ± 0.562 | 5.084 ± 0.703 |
2.307 ± 0.44 | 5.436 ± 0.556 |
6.453 ± 0.323 | 8.174 ± 0.601 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.62 ± 0.222 | 3.715 ± 0.487 |
5.28 ± 0.839 | 2.816 ± 0.411 |
5.984 ± 0.198 | 7.822 ± 0.344 |
6.023 ± 0.631 | 7.352 ± 0.493 |
1.525 ± 0.351 | 3.285 ± 0.315 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
