
Geodermatophilus sp. DF01_2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Geodermatophilus; unclassified Geodermatophilus
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
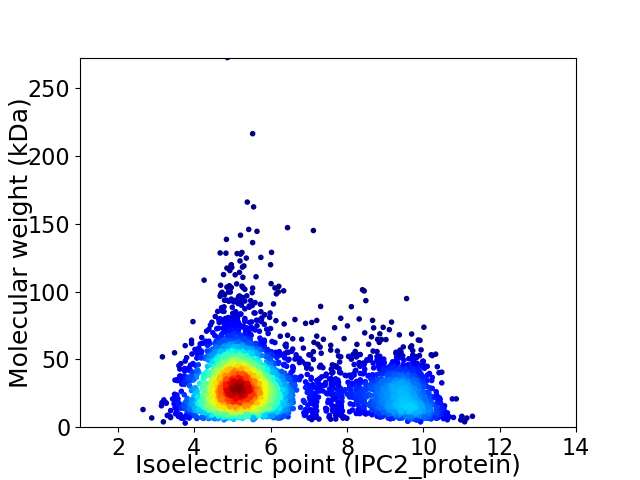
Virtual 2D-PAGE plot for 4069 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9N948|A0A4Y9N948_9ACTN Phosphoenolpyruvate carboxykinase [GTP] OS=Geodermatophilus sp. DF01_2 OX=2559610 GN=pckG PE=3 SV=1
MM1 pKa = 7.35RR2 pKa = 11.84RR3 pKa = 11.84AAVALLATAALLLVSGCSVVVVGRR27 pKa = 11.84PSAAQPPAADD37 pKa = 3.66VPPGEE42 pKa = 4.4VGVAGATEE50 pKa = 4.07EE51 pKa = 4.26PVDD54 pKa = 3.78VLARR58 pKa = 11.84NALADD63 pKa = 3.97LEE65 pKa = 4.92TYY67 pKa = 7.49WTEE70 pKa = 3.62QFPDD74 pKa = 3.27VFGQEE79 pKa = 4.18FVPLQGGYY87 pKa = 10.57FSVDD91 pKa = 3.36PDD93 pKa = 4.09DD94 pKa = 4.83VDD96 pKa = 3.67PAAYY100 pKa = 8.43PQGIGCGEE108 pKa = 4.4DD109 pKa = 3.48PRR111 pKa = 11.84DD112 pKa = 3.59VEE114 pKa = 5.52SNAFYY119 pKa = 10.67CQAPDD124 pKa = 3.76APNSDD129 pKa = 3.97SISYY133 pKa = 10.51DD134 pKa = 3.0RR135 pKa = 11.84AFLQEE140 pKa = 3.95LAEE143 pKa = 4.1QYY145 pKa = 11.33GRR147 pKa = 11.84FIPALVMAHH156 pKa = 6.2EE157 pKa = 5.39FGHH160 pKa = 6.49AVQGRR165 pKa = 11.84VGYY168 pKa = 9.0PDD170 pKa = 5.09DD171 pKa = 5.1SISVEE176 pKa = 4.11TQADD180 pKa = 4.0CFAGAWTAWVADD192 pKa = 3.98GQAEE196 pKa = 4.23HH197 pKa = 6.15SQIRR201 pKa = 11.84APEE204 pKa = 4.1LDD206 pKa = 3.48EE207 pKa = 4.08VLRR210 pKa = 11.84GYY212 pKa = 11.13LLLRR216 pKa = 11.84DD217 pKa = 3.88PVGVTTEE224 pKa = 4.11AAHH227 pKa = 6.46GSYY230 pKa = 10.46FDD232 pKa = 3.78RR233 pKa = 11.84VSAFQEE239 pKa = 4.48GFDD242 pKa = 4.93AGPTVCRR249 pKa = 11.84DD250 pKa = 2.93EE251 pKa = 4.6FTAQRR256 pKa = 11.84PYY258 pKa = 10.16TQDD261 pKa = 2.83AFSTANEE268 pKa = 4.01AASGGNAPFGLAQQYY283 pKa = 9.99AAEE286 pKa = 4.56ALPEE290 pKa = 3.91FWDD293 pKa = 3.64RR294 pKa = 11.84AFAAEE299 pKa = 4.09FGEE302 pKa = 4.7TFVSPTLEE310 pKa = 4.31PFAGTAPTCAPDD322 pKa = 4.7DD323 pKa = 4.23RR324 pKa = 11.84DD325 pKa = 3.98LVLCADD331 pKa = 4.5EE332 pKa = 5.77DD333 pKa = 4.24LVGYY337 pKa = 8.37DD338 pKa = 3.26VRR340 pKa = 11.84DD341 pKa = 3.59LVGPAYY347 pKa = 9.58EE348 pKa = 4.1QLEE351 pKa = 4.43SADD354 pKa = 3.72YY355 pKa = 11.12AVITAASLPYY365 pKa = 10.54ALAARR370 pKa = 11.84EE371 pKa = 3.87QLGLSTDD378 pKa = 3.6DD379 pKa = 3.33EE380 pKa = 4.42AAIRR384 pKa = 11.84SAVCLTGWFSRR395 pKa = 11.84AFFDD399 pKa = 5.53GEE401 pKa = 4.42LEE403 pKa = 4.2AADD406 pKa = 4.51ISPGDD411 pKa = 3.39IDD413 pKa = 4.02EE414 pKa = 4.95AVQFLLTYY422 pKa = 8.74GTDD425 pKa = 3.37PSVVPEE431 pKa = 4.06VEE433 pKa = 3.88LTGFQLVDD441 pKa = 3.55LFRR444 pKa = 11.84NGFFDD449 pKa = 4.68GAAACDD455 pKa = 3.55VGLL458 pKa = 4.5
MM1 pKa = 7.35RR2 pKa = 11.84RR3 pKa = 11.84AAVALLATAALLLVSGCSVVVVGRR27 pKa = 11.84PSAAQPPAADD37 pKa = 3.66VPPGEE42 pKa = 4.4VGVAGATEE50 pKa = 4.07EE51 pKa = 4.26PVDD54 pKa = 3.78VLARR58 pKa = 11.84NALADD63 pKa = 3.97LEE65 pKa = 4.92TYY67 pKa = 7.49WTEE70 pKa = 3.62QFPDD74 pKa = 3.27VFGQEE79 pKa = 4.18FVPLQGGYY87 pKa = 10.57FSVDD91 pKa = 3.36PDD93 pKa = 4.09DD94 pKa = 4.83VDD96 pKa = 3.67PAAYY100 pKa = 8.43PQGIGCGEE108 pKa = 4.4DD109 pKa = 3.48PRR111 pKa = 11.84DD112 pKa = 3.59VEE114 pKa = 5.52SNAFYY119 pKa = 10.67CQAPDD124 pKa = 3.76APNSDD129 pKa = 3.97SISYY133 pKa = 10.51DD134 pKa = 3.0RR135 pKa = 11.84AFLQEE140 pKa = 3.95LAEE143 pKa = 4.1QYY145 pKa = 11.33GRR147 pKa = 11.84FIPALVMAHH156 pKa = 6.2EE157 pKa = 5.39FGHH160 pKa = 6.49AVQGRR165 pKa = 11.84VGYY168 pKa = 9.0PDD170 pKa = 5.09DD171 pKa = 5.1SISVEE176 pKa = 4.11TQADD180 pKa = 4.0CFAGAWTAWVADD192 pKa = 3.98GQAEE196 pKa = 4.23HH197 pKa = 6.15SQIRR201 pKa = 11.84APEE204 pKa = 4.1LDD206 pKa = 3.48EE207 pKa = 4.08VLRR210 pKa = 11.84GYY212 pKa = 11.13LLLRR216 pKa = 11.84DD217 pKa = 3.88PVGVTTEE224 pKa = 4.11AAHH227 pKa = 6.46GSYY230 pKa = 10.46FDD232 pKa = 3.78RR233 pKa = 11.84VSAFQEE239 pKa = 4.48GFDD242 pKa = 4.93AGPTVCRR249 pKa = 11.84DD250 pKa = 2.93EE251 pKa = 4.6FTAQRR256 pKa = 11.84PYY258 pKa = 10.16TQDD261 pKa = 2.83AFSTANEE268 pKa = 4.01AASGGNAPFGLAQQYY283 pKa = 9.99AAEE286 pKa = 4.56ALPEE290 pKa = 3.91FWDD293 pKa = 3.64RR294 pKa = 11.84AFAAEE299 pKa = 4.09FGEE302 pKa = 4.7TFVSPTLEE310 pKa = 4.31PFAGTAPTCAPDD322 pKa = 4.7DD323 pKa = 4.23RR324 pKa = 11.84DD325 pKa = 3.98LVLCADD331 pKa = 4.5EE332 pKa = 5.77DD333 pKa = 4.24LVGYY337 pKa = 8.37DD338 pKa = 3.26VRR340 pKa = 11.84DD341 pKa = 3.59LVGPAYY347 pKa = 9.58EE348 pKa = 4.1QLEE351 pKa = 4.43SADD354 pKa = 3.72YY355 pKa = 11.12AVITAASLPYY365 pKa = 10.54ALAARR370 pKa = 11.84EE371 pKa = 3.87QLGLSTDD378 pKa = 3.6DD379 pKa = 3.33EE380 pKa = 4.42AAIRR384 pKa = 11.84SAVCLTGWFSRR395 pKa = 11.84AFFDD399 pKa = 5.53GEE401 pKa = 4.42LEE403 pKa = 4.2AADD406 pKa = 4.51ISPGDD411 pKa = 3.39IDD413 pKa = 4.02EE414 pKa = 4.95AVQFLLTYY422 pKa = 8.74GTDD425 pKa = 3.37PSVVPEE431 pKa = 4.06VEE433 pKa = 3.88LTGFQLVDD441 pKa = 3.55LFRR444 pKa = 11.84NGFFDD449 pKa = 4.68GAAACDD455 pKa = 3.55VGLL458 pKa = 4.5
Molecular weight: 48.82 kDa
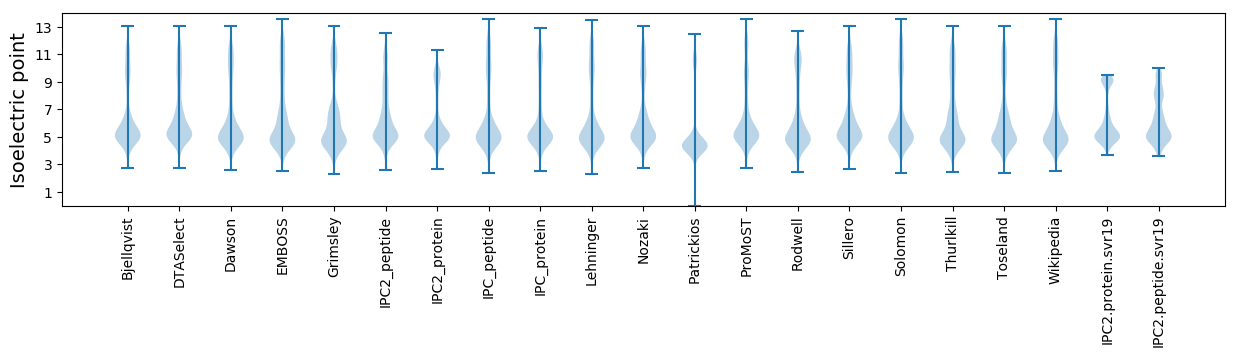
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9MHB7|A0A4Y9MHB7_9ACTN GAF domain-containing protein OS=Geodermatophilus sp. DF01_2 OX=2559610 GN=E4P41_18625 PE=4 SV=1
RR1 pKa = 7.57PGAPGAAVRR10 pKa = 11.84RR11 pKa = 11.84PPLAPRR17 pKa = 11.84PAAGRR22 pKa = 11.84PRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84PRR29 pKa = 11.84SAGRR33 pKa = 11.84PRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84PAAGSRR43 pKa = 11.84RR44 pKa = 11.84PARR47 pKa = 11.84RR48 pKa = 11.84SAPARR53 pKa = 11.84ARR55 pKa = 11.84RR56 pKa = 11.84ASRR59 pKa = 11.84PAAGGRR65 pKa = 11.84APAPPSRR72 pKa = 11.84CRR74 pKa = 11.84SGG76 pKa = 3.28
RR1 pKa = 7.57PGAPGAAVRR10 pKa = 11.84RR11 pKa = 11.84PPLAPRR17 pKa = 11.84PAAGRR22 pKa = 11.84PRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84PRR29 pKa = 11.84SAGRR33 pKa = 11.84PRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84PAAGSRR43 pKa = 11.84RR44 pKa = 11.84PARR47 pKa = 11.84RR48 pKa = 11.84SAPARR53 pKa = 11.84ARR55 pKa = 11.84RR56 pKa = 11.84ASRR59 pKa = 11.84PAAGGRR65 pKa = 11.84APAPPSRR72 pKa = 11.84CRR74 pKa = 11.84SGG76 pKa = 3.28
Molecular weight: 8.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1250318 |
32 |
2622 |
307.3 |
32.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.307 ± 0.062 | 0.721 ± 0.009 |
6.25 ± 0.03 | 5.781 ± 0.039 |
2.487 ± 0.021 | 9.66 ± 0.038 |
2.048 ± 0.018 | 2.673 ± 0.027 |
1.311 ± 0.021 | 10.628 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.588 ± 0.014 | 1.393 ± 0.016 |
6.329 ± 0.031 | 2.697 ± 0.019 |
8.542 ± 0.042 | 4.688 ± 0.027 |
5.839 ± 0.026 | 9.87 ± 0.036 |
1.444 ± 0.017 | 1.744 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
