
Marivirga lumbricoides
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Marivirgaceae; Marivirga
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
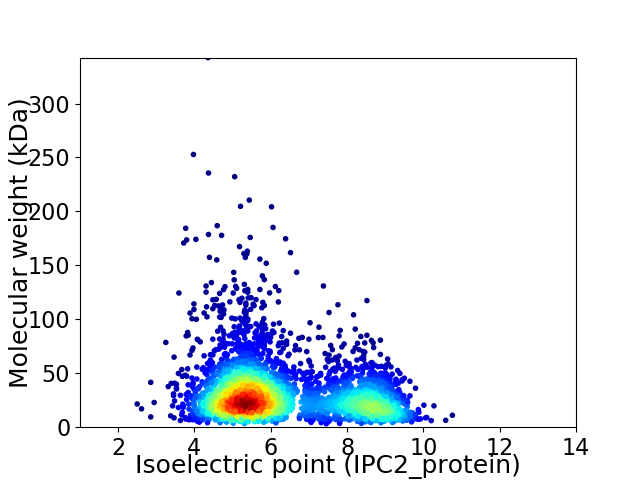
Virtual 2D-PAGE plot for 3452 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T4DHK9|A0A2T4DHK9_9BACT Uncharacterized protein OS=Marivirga lumbricoides OX=1046115 GN=C9994_13130 PE=4 SV=1
QQ1 pKa = 8.12DD2 pKa = 5.71LLPQHH7 pKa = 6.49NLPLHH12 pKa = 5.73YY13 pKa = 10.6LLLPQTTAFNVTVTDD28 pKa = 3.98GAGTEE33 pKa = 4.53LSFSPVTVSRR43 pKa = 11.84TAPAIIPLSSGAGSPKK59 pKa = 10.41GPNTKK64 pKa = 10.14FLVDD68 pKa = 4.38DD69 pKa = 4.06SQLATVQTDD78 pKa = 2.9EE79 pKa = 5.02GLILTANKK87 pKa = 10.58AFFTSIRR94 pKa = 11.84ISEE97 pKa = 4.25SAQAGFLTSKK107 pKa = 9.15GTAGFGKK114 pKa = 9.85EE115 pKa = 4.03FRR117 pKa = 11.84TGHH120 pKa = 5.13IWNTTGAAGRR130 pKa = 11.84KK131 pKa = 8.05AHH133 pKa = 6.05VFSFMATEE141 pKa = 4.48DD142 pKa = 3.5ATTVTISDD150 pKa = 4.16FGAVDD155 pKa = 3.95FEE157 pKa = 5.08NISEE161 pKa = 4.27SGDD164 pKa = 3.11ITVSLDD170 pKa = 3.24AGDD173 pKa = 4.51SYY175 pKa = 12.07VMAAFVDD182 pKa = 4.44KK183 pKa = 10.86PSTDD187 pKa = 2.94NLNNVNGTRR196 pKa = 11.84ISSNKK201 pKa = 9.84NIVVNSGSWLGGSPGGTDD219 pKa = 3.18EE220 pKa = 5.18GRR222 pKa = 11.84DD223 pKa = 3.18IGLDD227 pKa = 3.12QIAPIEE233 pKa = 4.22QTGFEE238 pKa = 4.39YY239 pKa = 10.47ILVKK243 pKa = 10.6GQGIASEE250 pKa = 4.12NVIVVAHH257 pKa = 6.52TDD259 pKa = 3.2GTDD262 pKa = 2.68IFLNGASSPANASPLNAGEE281 pKa = 3.91FHH283 pKa = 7.31RR284 pKa = 11.84FTASDD289 pKa = 3.65YY290 pKa = 10.81SANEE294 pKa = 3.53NMYY297 pKa = 8.64ITSNQPVYY305 pKa = 10.73VYY307 pKa = 10.6QGLNGANSTNEE318 pKa = 3.75RR319 pKa = 11.84QLGMNYY325 pKa = 8.5MPPIICLGGTNVDD338 pKa = 3.58IPNINQLGNPVIQIIAEE355 pKa = 4.11AGEE358 pKa = 4.24QVTITDD364 pKa = 3.79GSGTTDD370 pKa = 3.07VTSSARR376 pKa = 11.84AVTGNSNYY384 pKa = 7.83VTYY387 pKa = 11.03KK388 pKa = 10.04LAGYY392 pKa = 7.32TGNVTVEE399 pKa = 4.09SPRR402 pKa = 11.84PIRR405 pKa = 11.84VSLTIEE411 pKa = 3.86SGNIGGAGFFSGFTTSPVIEE431 pKa = 4.49TPSGYY436 pKa = 10.34DD437 pKa = 3.12ATSCIPDD444 pKa = 3.89NLPVTLTATGFDD456 pKa = 3.49SYY458 pKa = 10.23QWYY461 pKa = 10.03RR462 pKa = 11.84DD463 pKa = 3.82GIILTGEE470 pKa = 4.13TNASVSVDD478 pKa = 3.06KK479 pKa = 11.11PGVYY483 pKa = 8.66TAAGTIAGCVSSEE496 pKa = 3.7QSFPLSVTLCPGDD509 pKa = 3.91LGVAKK514 pKa = 10.53DD515 pKa = 3.94VVSVTNVSGDD525 pKa = 3.42LFDD528 pKa = 4.62VVFDD532 pKa = 5.49LIVTNYY538 pKa = 10.12SPSNPAPNMQLIDD551 pKa = 5.68DD552 pKa = 4.41ITDD555 pKa = 4.19GLPSGATASIQSAPTVVNGSFTTGGISSSYY585 pKa = 11.04NGTSDD590 pKa = 3.19IALLTTSLSGADD602 pKa = 3.66TEE604 pKa = 4.5LAAASTVTVRR614 pKa = 11.84FKK616 pKa = 11.38VRR618 pKa = 11.84VDD620 pKa = 3.17MSAATSPAYY629 pKa = 10.1AGQAVVTTAISGPNDD644 pKa = 4.68GITTTFDD651 pKa = 3.23NQDD654 pKa = 3.36FSDD657 pKa = 3.99SGSNPDD663 pKa = 3.83PNGNGDD669 pKa = 3.49PTEE672 pKa = 4.37AGEE675 pKa = 3.98NDD677 pKa = 3.0ITEE680 pKa = 4.36VCLSNTTISYY690 pKa = 10.03SASTYY695 pKa = 10.52YY696 pKa = 9.85STGSDD701 pKa = 3.48PTPIINGVSGGTFSAPKK718 pKa = 10.13GLVINSTSGEE728 pKa = 4.0IDD730 pKa = 3.24VSASIVDD737 pKa = 4.17DD738 pKa = 4.51YY739 pKa = 11.63IVTYY743 pKa = 10.64SFGGLCPSTTSISIALNPPAEE764 pKa = 4.14PTVINQITNNTTPTLTGTAVVNVGEE789 pKa = 4.22SLEE792 pKa = 4.05VLLNGQTYY800 pKa = 10.7VLGDD804 pKa = 3.78GNLSISGTTWTLIVPVANAIGTDD827 pKa = 4.38GIYY830 pKa = 10.8DD831 pKa = 3.48VTATHH836 pKa = 6.66INGVGTRR843 pKa = 11.84TDD845 pKa = 3.26DD846 pKa = 3.53TTNGEE851 pKa = 4.25LEE853 pKa = 4.33IDD855 pKa = 3.37TDD857 pKa = 3.63APEE860 pKa = 4.3IVISGQPSIINNNSSYY876 pKa = 10.41TVNLNFDD883 pKa = 3.58EE884 pKa = 6.33DD885 pKa = 3.71ITGLQITDD893 pKa = 3.15IVVTNASLSNLTSINASTYY912 pKa = 9.6TIVVLPAGTGNITIDD927 pKa = 4.08FPAGGVKK934 pKa = 10.4DD935 pKa = 3.57LANNNNLAASQVLTIYY951 pKa = 10.99DD952 pKa = 3.79DD953 pKa = 3.83TAPATPTVNALTTSDD968 pKa = 3.62NTPVVSGTAEE978 pKa = 3.72IGSTVTVIINGVTFTTTADD997 pKa = 3.33ASGNWSVDD1005 pKa = 3.14TEE1007 pKa = 4.6TATPTAGGPFTPLTDD1022 pKa = 3.16GDD1024 pKa = 3.95YY1025 pKa = 11.32DD1026 pKa = 3.96VAVTSTDD1033 pKa = 3.11AAGNSSSDD1041 pKa = 3.17ATSNEE1046 pKa = 3.55VSIDD1050 pKa = 3.46TTAPTAPIVNLLTTNDD1066 pKa = 3.71VTPVITGTNGLGTSQLAGEE1085 pKa = 4.46TLTVTVNGATYY1096 pKa = 9.57TVTPDD1101 pKa = 3.57ASGNWSVDD1109 pKa = 3.14TEE1111 pKa = 4.57TATPTSGTLGTFNDD1125 pKa = 4.13GTSYY1129 pKa = 11.01EE1130 pKa = 4.39VVATVTDD1137 pKa = 3.68AASNSTSDD1145 pKa = 3.14ATSNEE1150 pKa = 3.67VSIDD1154 pKa = 3.36TTAPATPTVNTLTTSDD1170 pKa = 3.85NTPVLSGTAEE1180 pKa = 3.77IGSTVVVVINGVTFTTTADD1199 pKa = 3.43ASGDD1203 pKa = 3.28WSVDD1207 pKa = 3.17TEE1209 pKa = 4.57TATPTAGG1216 pKa = 3.2
QQ1 pKa = 8.12DD2 pKa = 5.71LLPQHH7 pKa = 6.49NLPLHH12 pKa = 5.73YY13 pKa = 10.6LLLPQTTAFNVTVTDD28 pKa = 3.98GAGTEE33 pKa = 4.53LSFSPVTVSRR43 pKa = 11.84TAPAIIPLSSGAGSPKK59 pKa = 10.41GPNTKK64 pKa = 10.14FLVDD68 pKa = 4.38DD69 pKa = 4.06SQLATVQTDD78 pKa = 2.9EE79 pKa = 5.02GLILTANKK87 pKa = 10.58AFFTSIRR94 pKa = 11.84ISEE97 pKa = 4.25SAQAGFLTSKK107 pKa = 9.15GTAGFGKK114 pKa = 9.85EE115 pKa = 4.03FRR117 pKa = 11.84TGHH120 pKa = 5.13IWNTTGAAGRR130 pKa = 11.84KK131 pKa = 8.05AHH133 pKa = 6.05VFSFMATEE141 pKa = 4.48DD142 pKa = 3.5ATTVTISDD150 pKa = 4.16FGAVDD155 pKa = 3.95FEE157 pKa = 5.08NISEE161 pKa = 4.27SGDD164 pKa = 3.11ITVSLDD170 pKa = 3.24AGDD173 pKa = 4.51SYY175 pKa = 12.07VMAAFVDD182 pKa = 4.44KK183 pKa = 10.86PSTDD187 pKa = 2.94NLNNVNGTRR196 pKa = 11.84ISSNKK201 pKa = 9.84NIVVNSGSWLGGSPGGTDD219 pKa = 3.18EE220 pKa = 5.18GRR222 pKa = 11.84DD223 pKa = 3.18IGLDD227 pKa = 3.12QIAPIEE233 pKa = 4.22QTGFEE238 pKa = 4.39YY239 pKa = 10.47ILVKK243 pKa = 10.6GQGIASEE250 pKa = 4.12NVIVVAHH257 pKa = 6.52TDD259 pKa = 3.2GTDD262 pKa = 2.68IFLNGASSPANASPLNAGEE281 pKa = 3.91FHH283 pKa = 7.31RR284 pKa = 11.84FTASDD289 pKa = 3.65YY290 pKa = 10.81SANEE294 pKa = 3.53NMYY297 pKa = 8.64ITSNQPVYY305 pKa = 10.73VYY307 pKa = 10.6QGLNGANSTNEE318 pKa = 3.75RR319 pKa = 11.84QLGMNYY325 pKa = 8.5MPPIICLGGTNVDD338 pKa = 3.58IPNINQLGNPVIQIIAEE355 pKa = 4.11AGEE358 pKa = 4.24QVTITDD364 pKa = 3.79GSGTTDD370 pKa = 3.07VTSSARR376 pKa = 11.84AVTGNSNYY384 pKa = 7.83VTYY387 pKa = 11.03KK388 pKa = 10.04LAGYY392 pKa = 7.32TGNVTVEE399 pKa = 4.09SPRR402 pKa = 11.84PIRR405 pKa = 11.84VSLTIEE411 pKa = 3.86SGNIGGAGFFSGFTTSPVIEE431 pKa = 4.49TPSGYY436 pKa = 10.34DD437 pKa = 3.12ATSCIPDD444 pKa = 3.89NLPVTLTATGFDD456 pKa = 3.49SYY458 pKa = 10.23QWYY461 pKa = 10.03RR462 pKa = 11.84DD463 pKa = 3.82GIILTGEE470 pKa = 4.13TNASVSVDD478 pKa = 3.06KK479 pKa = 11.11PGVYY483 pKa = 8.66TAAGTIAGCVSSEE496 pKa = 3.7QSFPLSVTLCPGDD509 pKa = 3.91LGVAKK514 pKa = 10.53DD515 pKa = 3.94VVSVTNVSGDD525 pKa = 3.42LFDD528 pKa = 4.62VVFDD532 pKa = 5.49LIVTNYY538 pKa = 10.12SPSNPAPNMQLIDD551 pKa = 5.68DD552 pKa = 4.41ITDD555 pKa = 4.19GLPSGATASIQSAPTVVNGSFTTGGISSSYY585 pKa = 11.04NGTSDD590 pKa = 3.19IALLTTSLSGADD602 pKa = 3.66TEE604 pKa = 4.5LAAASTVTVRR614 pKa = 11.84FKK616 pKa = 11.38VRR618 pKa = 11.84VDD620 pKa = 3.17MSAATSPAYY629 pKa = 10.1AGQAVVTTAISGPNDD644 pKa = 4.68GITTTFDD651 pKa = 3.23NQDD654 pKa = 3.36FSDD657 pKa = 3.99SGSNPDD663 pKa = 3.83PNGNGDD669 pKa = 3.49PTEE672 pKa = 4.37AGEE675 pKa = 3.98NDD677 pKa = 3.0ITEE680 pKa = 4.36VCLSNTTISYY690 pKa = 10.03SASTYY695 pKa = 10.52YY696 pKa = 9.85STGSDD701 pKa = 3.48PTPIINGVSGGTFSAPKK718 pKa = 10.13GLVINSTSGEE728 pKa = 4.0IDD730 pKa = 3.24VSASIVDD737 pKa = 4.17DD738 pKa = 4.51YY739 pKa = 11.63IVTYY743 pKa = 10.64SFGGLCPSTTSISIALNPPAEE764 pKa = 4.14PTVINQITNNTTPTLTGTAVVNVGEE789 pKa = 4.22SLEE792 pKa = 4.05VLLNGQTYY800 pKa = 10.7VLGDD804 pKa = 3.78GNLSISGTTWTLIVPVANAIGTDD827 pKa = 4.38GIYY830 pKa = 10.8DD831 pKa = 3.48VTATHH836 pKa = 6.66INGVGTRR843 pKa = 11.84TDD845 pKa = 3.26DD846 pKa = 3.53TTNGEE851 pKa = 4.25LEE853 pKa = 4.33IDD855 pKa = 3.37TDD857 pKa = 3.63APEE860 pKa = 4.3IVISGQPSIINNNSSYY876 pKa = 10.41TVNLNFDD883 pKa = 3.58EE884 pKa = 6.33DD885 pKa = 3.71ITGLQITDD893 pKa = 3.15IVVTNASLSNLTSINASTYY912 pKa = 9.6TIVVLPAGTGNITIDD927 pKa = 4.08FPAGGVKK934 pKa = 10.4DD935 pKa = 3.57LANNNNLAASQVLTIYY951 pKa = 10.99DD952 pKa = 3.79DD953 pKa = 3.83TAPATPTVNALTTSDD968 pKa = 3.62NTPVVSGTAEE978 pKa = 3.72IGSTVTVIINGVTFTTTADD997 pKa = 3.33ASGNWSVDD1005 pKa = 3.14TEE1007 pKa = 4.6TATPTAGGPFTPLTDD1022 pKa = 3.16GDD1024 pKa = 3.95YY1025 pKa = 11.32DD1026 pKa = 3.96VAVTSTDD1033 pKa = 3.11AAGNSSSDD1041 pKa = 3.17ATSNEE1046 pKa = 3.55VSIDD1050 pKa = 3.46TTAPTAPIVNLLTTNDD1066 pKa = 3.71VTPVITGTNGLGTSQLAGEE1085 pKa = 4.46TLTVTVNGATYY1096 pKa = 9.57TVTPDD1101 pKa = 3.57ASGNWSVDD1109 pKa = 3.14TEE1111 pKa = 4.57TATPTSGTLGTFNDD1125 pKa = 4.13GTSYY1129 pKa = 11.01EE1130 pKa = 4.39VVATVTDD1137 pKa = 3.68AASNSTSDD1145 pKa = 3.14ATSNEE1150 pKa = 3.67VSIDD1154 pKa = 3.36TTAPATPTVNTLTTSDD1170 pKa = 3.85NTPVLSGTAEE1180 pKa = 3.77IGSTVVVVINGVTFTTTADD1199 pKa = 3.43ASGDD1203 pKa = 3.28WSVDD1207 pKa = 3.17TEE1209 pKa = 4.57TATPTAGG1216 pKa = 3.2
Molecular weight: 124.36 kDa
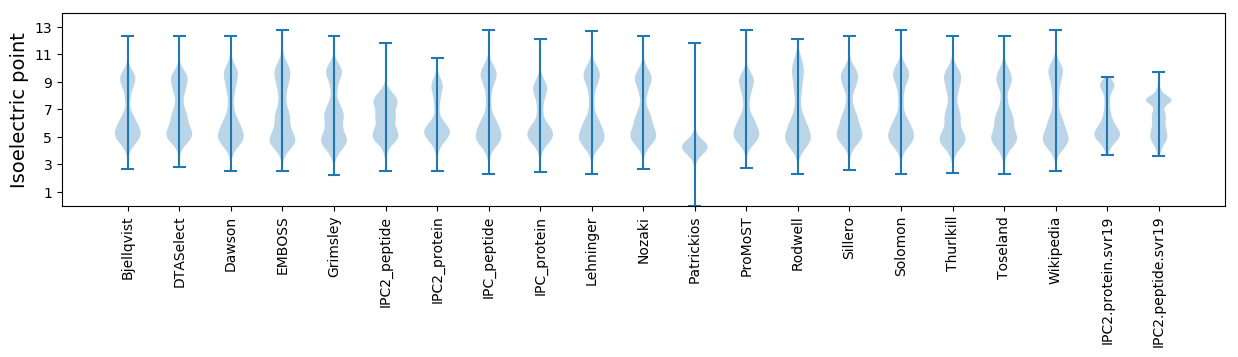
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T4DSC5|A0A2T4DSC5_9BACT SanA protein OS=Marivirga lumbricoides OX=1046115 GN=C9994_06145 PE=4 SV=1
MM1 pKa = 7.59ISALTNCRR9 pKa = 11.84KK10 pKa = 9.69ALKK13 pKa = 10.2YY14 pKa = 10.1YY15 pKa = 9.87SAWTTTLLGCRR26 pKa = 11.84WRR28 pKa = 11.84TVIKK32 pKa = 8.86TIPAAKK38 pKa = 8.9TVTGTMPVAGTTTAPGITRR57 pKa = 11.84CTTSIMMPLWVGLMWWILMPMSLPII82 pKa = 4.46
MM1 pKa = 7.59ISALTNCRR9 pKa = 11.84KK10 pKa = 9.69ALKK13 pKa = 10.2YY14 pKa = 10.1YY15 pKa = 9.87SAWTTTLLGCRR26 pKa = 11.84WRR28 pKa = 11.84TVIKK32 pKa = 8.86TIPAAKK38 pKa = 8.9TVTGTMPVAGTTTAPGITRR57 pKa = 11.84CTTSIMMPLWVGLMWWILMPMSLPII82 pKa = 4.46
Molecular weight: 9.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1013358 |
31 |
3123 |
293.6 |
33.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.448 ± 0.047 | 0.711 ± 0.013 |
5.442 ± 0.034 | 7.152 ± 0.044 |
5.208 ± 0.033 | 6.351 ± 0.044 |
1.774 ± 0.021 | 7.757 ± 0.036 |
7.348 ± 0.057 | 9.599 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.191 ± 0.02 | 5.89 ± 0.041 |
3.436 ± 0.026 | 3.716 ± 0.026 |
3.609 ± 0.028 | 6.895 ± 0.041 |
5.254 ± 0.044 | 5.986 ± 0.033 |
1.128 ± 0.015 | 4.104 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
