
Halorubrum sp. JWXQ-INN 858
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halorubrum; unclassified Halorubrum
Average proteome isoelectric point is 4.9
Get precalculated fractions of proteins
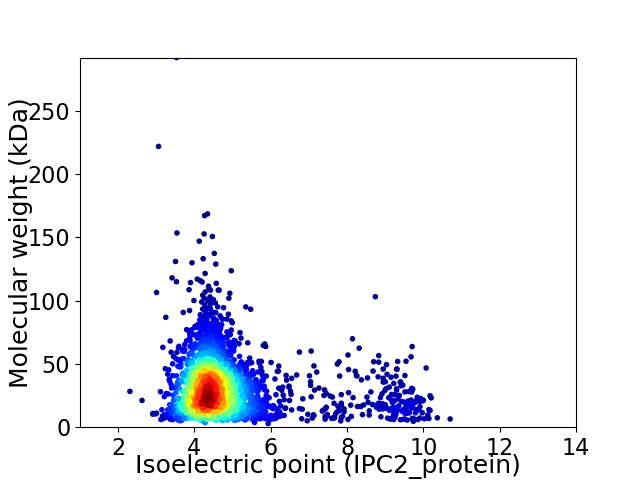
Virtual 2D-PAGE plot for 2889 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
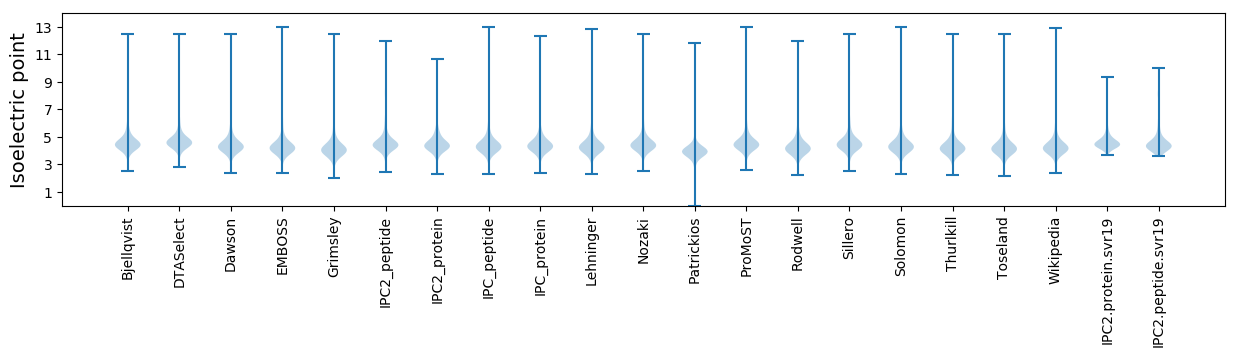
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G2HF61|A0A6G2HF61_9EURY Uncharacterized protein OS=Halorubrum sp. JWXQ-INN 858 OX=2690782 GN=GRS48_07690 PE=4 SV=1
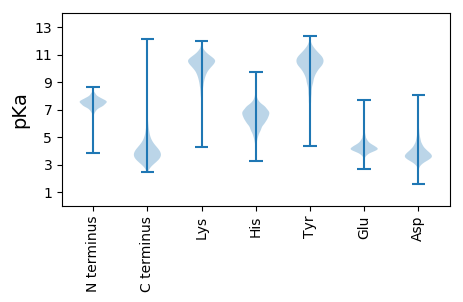
MM1 pKa = 7.64NDD3 pKa = 2.9TTRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FLVGTTAAGVAALAGCADD26 pKa = 4.78DD27 pKa = 5.18EE28 pKa = 5.69DD29 pKa = 5.41PDD31 pKa = 4.09EE32 pKa = 4.83TPADD36 pKa = 4.01APEE39 pKa = 5.28DD40 pKa = 4.31DD41 pKa = 4.4PDD43 pKa = 4.07EE44 pKa = 4.47PTEE47 pKa = 4.25AADD50 pKa = 5.08DD51 pKa = 4.3APADD55 pKa = 4.05DD56 pKa = 5.37EE57 pKa = 4.46PAEE60 pKa = 4.16SEE62 pKa = 3.92PTYY65 pKa = 9.87EE66 pKa = 4.72IWGADD71 pKa = 3.0QGTNTIYY78 pKa = 10.36IHH80 pKa = 7.25RR81 pKa = 11.84PTDD84 pKa = 3.64DD85 pKa = 3.67GGFEE89 pKa = 4.13QVDD92 pKa = 4.43EE93 pKa = 4.7IDD95 pKa = 3.31TGAVGGEE102 pKa = 4.13VPHH105 pKa = 6.1MVDD108 pKa = 3.05YY109 pKa = 10.75TSDD112 pKa = 3.48YY113 pKa = 10.35EE114 pKa = 4.34YY115 pKa = 11.31AAVACTVGGRR125 pKa = 11.84TLIYY129 pKa = 9.18RR130 pKa = 11.84TEE132 pKa = 3.98DD133 pKa = 3.32RR134 pKa = 11.84EE135 pKa = 4.2LVANLEE141 pKa = 4.42TGAGSHH147 pKa = 6.4FAGFSPDD154 pKa = 3.5DD155 pKa = 4.12EE156 pKa = 4.64YY157 pKa = 12.16VHH159 pKa = 7.58VDD161 pKa = 3.4VIGEE165 pKa = 4.07EE166 pKa = 4.09KK167 pKa = 10.06IVRR170 pKa = 11.84IDD172 pKa = 3.54ADD174 pKa = 3.91LEE176 pKa = 4.3NEE178 pKa = 4.13TFEE181 pKa = 4.22QVAEE185 pKa = 3.87IDD187 pKa = 3.6VRR189 pKa = 11.84EE190 pKa = 4.61DD191 pKa = 3.2EE192 pKa = 4.84TIVDD196 pKa = 3.92AGIEE200 pKa = 4.17TGSPICHH207 pKa = 6.34QYY209 pKa = 10.47AQDD212 pKa = 3.42GRR214 pKa = 11.84SLHH217 pKa = 5.63TLGPSYY223 pKa = 10.9HH224 pKa = 7.23DD225 pKa = 3.68AALVIVDD232 pKa = 4.6HH233 pKa = 7.88DD234 pKa = 4.25DD235 pKa = 3.84FSVDD239 pKa = 2.76RR240 pKa = 11.84AYY242 pKa = 10.02PGEE245 pKa = 4.23EE246 pKa = 3.98VPTNCGTVSHH256 pKa = 6.75PTDD259 pKa = 3.03EE260 pKa = 4.62KK261 pKa = 11.05FYY263 pKa = 9.94LTAGLPSDD271 pKa = 4.35PDD273 pKa = 3.65ADD275 pKa = 3.77PEE277 pKa = 4.28EE278 pKa = 4.55EE279 pKa = 4.25PEE281 pKa = 3.99AAAGVGDD288 pKa = 4.15FYY290 pKa = 11.75VYY292 pKa = 9.64DD293 pKa = 3.78TADD296 pKa = 3.85DD297 pKa = 3.83EE298 pKa = 4.83VATVGSTGGVDD309 pKa = 3.51AHH311 pKa = 6.42GFWFTPDD318 pKa = 4.14GEE320 pKa = 4.56EE321 pKa = 3.7LWVLNRR327 pKa = 11.84EE328 pKa = 4.32TNDD331 pKa = 3.92GVILDD336 pKa = 4.44PGTDD340 pKa = 3.33EE341 pKa = 4.97VVDD344 pKa = 4.93EE345 pKa = 4.67IDD347 pKa = 3.43AFGPEE352 pKa = 4.0QHH354 pKa = 7.47DD355 pKa = 4.71DD356 pKa = 3.78PTEE359 pKa = 4.3SDD361 pKa = 4.04APDD364 pKa = 3.65IMWASPDD371 pKa = 3.53GEE373 pKa = 4.65YY374 pKa = 10.36MFVTLRR380 pKa = 11.84GPNPLSGDD388 pKa = 3.32PHH390 pKa = 6.9AATGINPGYY399 pKa = 10.88AVFDD403 pKa = 3.7VEE405 pKa = 4.3SRR407 pKa = 11.84EE408 pKa = 4.24RR409 pKa = 11.84VDD411 pKa = 5.28LVQPDD416 pKa = 3.89GDD418 pKa = 4.03NPDD421 pKa = 3.65SDD423 pKa = 3.79FHH425 pKa = 8.23GIGVRR430 pKa = 11.84PLTDD434 pKa = 3.12HH435 pKa = 7.64DD436 pKa = 4.93GFTSPPYY443 pKa = 10.63
MM1 pKa = 7.64NDD3 pKa = 2.9TTRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FLVGTTAAGVAALAGCADD26 pKa = 4.78DD27 pKa = 5.18EE28 pKa = 5.69DD29 pKa = 5.41PDD31 pKa = 4.09EE32 pKa = 4.83TPADD36 pKa = 4.01APEE39 pKa = 5.28DD40 pKa = 4.31DD41 pKa = 4.4PDD43 pKa = 4.07EE44 pKa = 4.47PTEE47 pKa = 4.25AADD50 pKa = 5.08DD51 pKa = 4.3APADD55 pKa = 4.05DD56 pKa = 5.37EE57 pKa = 4.46PAEE60 pKa = 4.16SEE62 pKa = 3.92PTYY65 pKa = 9.87EE66 pKa = 4.72IWGADD71 pKa = 3.0QGTNTIYY78 pKa = 10.36IHH80 pKa = 7.25RR81 pKa = 11.84PTDD84 pKa = 3.64DD85 pKa = 3.67GGFEE89 pKa = 4.13QVDD92 pKa = 4.43EE93 pKa = 4.7IDD95 pKa = 3.31TGAVGGEE102 pKa = 4.13VPHH105 pKa = 6.1MVDD108 pKa = 3.05YY109 pKa = 10.75TSDD112 pKa = 3.48YY113 pKa = 10.35EE114 pKa = 4.34YY115 pKa = 11.31AAVACTVGGRR125 pKa = 11.84TLIYY129 pKa = 9.18RR130 pKa = 11.84TEE132 pKa = 3.98DD133 pKa = 3.32RR134 pKa = 11.84EE135 pKa = 4.2LVANLEE141 pKa = 4.42TGAGSHH147 pKa = 6.4FAGFSPDD154 pKa = 3.5DD155 pKa = 4.12EE156 pKa = 4.64YY157 pKa = 12.16VHH159 pKa = 7.58VDD161 pKa = 3.4VIGEE165 pKa = 4.07EE166 pKa = 4.09KK167 pKa = 10.06IVRR170 pKa = 11.84IDD172 pKa = 3.54ADD174 pKa = 3.91LEE176 pKa = 4.3NEE178 pKa = 4.13TFEE181 pKa = 4.22QVAEE185 pKa = 3.87IDD187 pKa = 3.6VRR189 pKa = 11.84EE190 pKa = 4.61DD191 pKa = 3.2EE192 pKa = 4.84TIVDD196 pKa = 3.92AGIEE200 pKa = 4.17TGSPICHH207 pKa = 6.34QYY209 pKa = 10.47AQDD212 pKa = 3.42GRR214 pKa = 11.84SLHH217 pKa = 5.63TLGPSYY223 pKa = 10.9HH224 pKa = 7.23DD225 pKa = 3.68AALVIVDD232 pKa = 4.6HH233 pKa = 7.88DD234 pKa = 4.25DD235 pKa = 3.84FSVDD239 pKa = 2.76RR240 pKa = 11.84AYY242 pKa = 10.02PGEE245 pKa = 4.23EE246 pKa = 3.98VPTNCGTVSHH256 pKa = 6.75PTDD259 pKa = 3.03EE260 pKa = 4.62KK261 pKa = 11.05FYY263 pKa = 9.94LTAGLPSDD271 pKa = 4.35PDD273 pKa = 3.65ADD275 pKa = 3.77PEE277 pKa = 4.28EE278 pKa = 4.55EE279 pKa = 4.25PEE281 pKa = 3.99AAAGVGDD288 pKa = 4.15FYY290 pKa = 11.75VYY292 pKa = 9.64DD293 pKa = 3.78TADD296 pKa = 3.85DD297 pKa = 3.83EE298 pKa = 4.83VATVGSTGGVDD309 pKa = 3.51AHH311 pKa = 6.42GFWFTPDD318 pKa = 4.14GEE320 pKa = 4.56EE321 pKa = 3.7LWVLNRR327 pKa = 11.84EE328 pKa = 4.32TNDD331 pKa = 3.92GVILDD336 pKa = 4.44PGTDD340 pKa = 3.33EE341 pKa = 4.97VVDD344 pKa = 4.93EE345 pKa = 4.67IDD347 pKa = 3.43AFGPEE352 pKa = 4.0QHH354 pKa = 7.47DD355 pKa = 4.71DD356 pKa = 3.78PTEE359 pKa = 4.3SDD361 pKa = 4.04APDD364 pKa = 3.65IMWASPDD371 pKa = 3.53GEE373 pKa = 4.65YY374 pKa = 10.36MFVTLRR380 pKa = 11.84GPNPLSGDD388 pKa = 3.32PHH390 pKa = 6.9AATGINPGYY399 pKa = 10.88AVFDD403 pKa = 3.7VEE405 pKa = 4.3SRR407 pKa = 11.84EE408 pKa = 4.24RR409 pKa = 11.84VDD411 pKa = 5.28LVQPDD416 pKa = 3.89GDD418 pKa = 4.03NPDD421 pKa = 3.65SDD423 pKa = 3.79FHH425 pKa = 8.23GIGVRR430 pKa = 11.84PLTDD434 pKa = 3.12HH435 pKa = 7.64DD436 pKa = 4.93GFTSPPYY443 pKa = 10.63
Molecular weight: 47.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G2HI32|A0A6G2HI32_9EURY Uncharacterized protein OS=Halorubrum sp. JWXQ-INN 858 OX=2690782 GN=GRS48_11730 PE=4 SV=1
MM1 pKa = 7.14ATCDD5 pKa = 3.29VCGEE9 pKa = 4.29YY10 pKa = 10.86EE11 pKa = 4.06NLPYY15 pKa = 10.07QCRR18 pKa = 11.84RR19 pKa = 11.84CGRR22 pKa = 11.84TFCGEE27 pKa = 3.37HH28 pKa = 6.74RR29 pKa = 11.84LPEE32 pKa = 4.09NHH34 pKa = 7.34DD35 pKa = 3.83CPGLRR40 pKa = 11.84GSNDD44 pKa = 3.14PGAVFEE50 pKa = 4.58STVGGVDD57 pKa = 2.85RR58 pKa = 11.84GGVATGTGRR67 pKa = 11.84SSDD70 pKa = 3.24AGIVDD75 pKa = 3.7RR76 pKa = 11.84VKK78 pKa = 10.7RR79 pKa = 11.84RR80 pKa = 11.84VDD82 pKa = 3.25RR83 pKa = 11.84EE84 pKa = 4.05TGRR87 pKa = 11.84GNVMSRR93 pKa = 11.84FRR95 pKa = 11.84GKK97 pKa = 8.89ATYY100 pKa = 10.61VLLAAMWFTLLGQWAALAVGGSALHH125 pKa = 6.06NRR127 pKa = 11.84LFVLEE132 pKa = 5.97SYY134 pKa = 11.01ALWQVWTWGTAVFAHH149 pKa = 7.04DD150 pKa = 4.64PGLLFHH156 pKa = 7.39IIGNSIVLFFFGPLVEE172 pKa = 4.26RR173 pKa = 11.84AVGSKK178 pKa = 10.3RR179 pKa = 11.84FVGFFLVAGVLAGLGHH195 pKa = 5.96VLFVLATGGVVGVLGASGATFAILGVLTVWRR226 pKa = 11.84PNMQILLFFVVPMKK240 pKa = 10.72LKK242 pKa = 10.16YY243 pKa = 8.95LTWGIAAVSAFLVVSTGAAGVGGIAHH269 pKa = 7.91LAHH272 pKa = 7.35LIGFAIGLAFGKK284 pKa = 10.58RR285 pKa = 11.84NVNLARR291 pKa = 11.84RR292 pKa = 11.84TGGVGGVRR300 pKa = 11.84ASGVGGPGGPRR311 pKa = 11.84GPGGPGGRR319 pKa = 11.84FF320 pKa = 3.04
MM1 pKa = 7.14ATCDD5 pKa = 3.29VCGEE9 pKa = 4.29YY10 pKa = 10.86EE11 pKa = 4.06NLPYY15 pKa = 10.07QCRR18 pKa = 11.84RR19 pKa = 11.84CGRR22 pKa = 11.84TFCGEE27 pKa = 3.37HH28 pKa = 6.74RR29 pKa = 11.84LPEE32 pKa = 4.09NHH34 pKa = 7.34DD35 pKa = 3.83CPGLRR40 pKa = 11.84GSNDD44 pKa = 3.14PGAVFEE50 pKa = 4.58STVGGVDD57 pKa = 2.85RR58 pKa = 11.84GGVATGTGRR67 pKa = 11.84SSDD70 pKa = 3.24AGIVDD75 pKa = 3.7RR76 pKa = 11.84VKK78 pKa = 10.7RR79 pKa = 11.84RR80 pKa = 11.84VDD82 pKa = 3.25RR83 pKa = 11.84EE84 pKa = 4.05TGRR87 pKa = 11.84GNVMSRR93 pKa = 11.84FRR95 pKa = 11.84GKK97 pKa = 8.89ATYY100 pKa = 10.61VLLAAMWFTLLGQWAALAVGGSALHH125 pKa = 6.06NRR127 pKa = 11.84LFVLEE132 pKa = 5.97SYY134 pKa = 11.01ALWQVWTWGTAVFAHH149 pKa = 7.04DD150 pKa = 4.64PGLLFHH156 pKa = 7.39IIGNSIVLFFFGPLVEE172 pKa = 4.26RR173 pKa = 11.84AVGSKK178 pKa = 10.3RR179 pKa = 11.84FVGFFLVAGVLAGLGHH195 pKa = 5.96VLFVLATGGVVGVLGASGATFAILGVLTVWRR226 pKa = 11.84PNMQILLFFVVPMKK240 pKa = 10.72LKK242 pKa = 10.16YY243 pKa = 8.95LTWGIAAVSAFLVVSTGAAGVGGIAHH269 pKa = 7.91LAHH272 pKa = 7.35LIGFAIGLAFGKK284 pKa = 10.58RR285 pKa = 11.84NVNLARR291 pKa = 11.84RR292 pKa = 11.84TGGVGGVRR300 pKa = 11.84ASGVGGPGGPRR311 pKa = 11.84GPGGPGGRR319 pKa = 11.84FF320 pKa = 3.04
Molecular weight: 33.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
872988 |
27 |
2830 |
302.2 |
32.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.108 ± 0.067 | 0.662 ± 0.015 |
9.07 ± 0.064 | 7.842 ± 0.061 |
3.216 ± 0.025 | 9.302 ± 0.053 |
1.961 ± 0.019 | 3.768 ± 0.034 |
1.57 ± 0.029 | 8.612 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.618 ± 0.021 | 2.044 ± 0.023 |
4.879 ± 0.028 | 1.796 ± 0.024 |
7.035 ± 0.048 | 4.999 ± 0.031 |
6.249 ± 0.041 | 9.717 ± 0.055 |
1.053 ± 0.015 | 2.499 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
