
Shewanella aestuarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Shewanellaceae; Shewanella
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
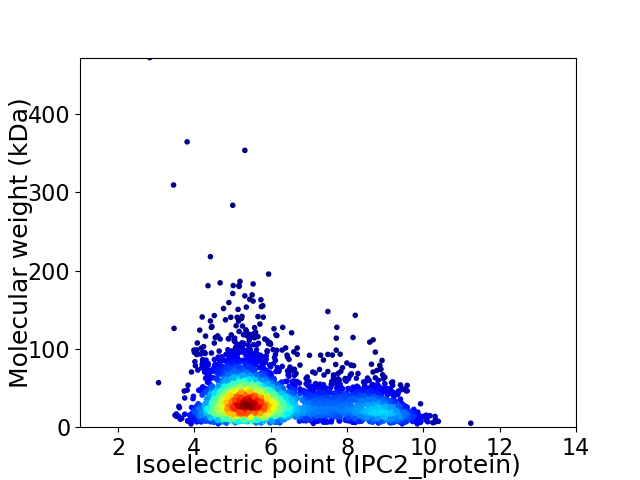
Virtual 2D-PAGE plot for 3605 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
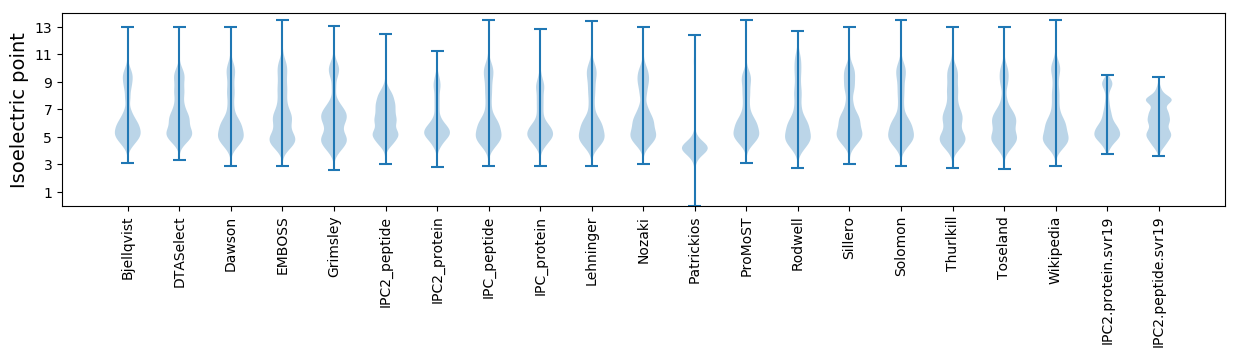
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G9QNB3|A0A6G9QNB3_9GAMM Na(+)-translocating NADH-quinone reductase subunit D OS=Shewanella aestuarii OX=1028752 GN=nqrD PE=3 SV=1
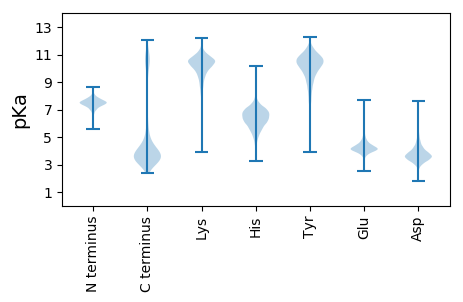
MM1 pKa = 7.34KK2 pKa = 10.22RR3 pKa = 11.84IYY5 pKa = 10.35PFALCISLVGLTACSDD21 pKa = 3.76DD22 pKa = 5.7DD23 pKa = 4.38DD24 pKa = 5.99DD25 pKa = 6.2KK26 pKa = 11.93VVTPDD31 pKa = 3.43TPEE34 pKa = 3.59VEE36 pKa = 4.4YY37 pKa = 11.07SHH39 pKa = 7.04VRR41 pKa = 11.84VIHH44 pKa = 6.39AGSDD48 pKa = 3.19APMVNVMANGAALLSDD64 pKa = 3.43VDD66 pKa = 4.02YY67 pKa = 11.77AMSSGLLEE75 pKa = 4.35VTSATYY81 pKa = 10.45DD82 pKa = 3.25IDD84 pKa = 4.1VDD86 pKa = 3.92ALLADD91 pKa = 4.21GTTLTVLEE99 pKa = 4.87ANLATEE105 pKa = 4.66ANTEE109 pKa = 4.11YY110 pKa = 10.31TAVALGTVADD120 pKa = 4.37EE121 pKa = 4.63SLMLKK126 pKa = 10.55LIANPTAEE134 pKa = 4.0IAAGYY139 pKa = 10.84ARR141 pKa = 11.84VQVLHH146 pKa = 6.2ATPSVGLVDD155 pKa = 4.21VYY157 pKa = 10.1VTAPGDD163 pKa = 4.44DD164 pKa = 3.39ISAIAPTLSANYY176 pKa = 8.8MDD178 pKa = 5.48NSTQLEE184 pKa = 4.71VPVGDD189 pKa = 3.54YY190 pKa = 10.46QIRR193 pKa = 11.84ITGSNSKK200 pKa = 10.33EE201 pKa = 3.88VVFDD205 pKa = 3.61SGTVALGDD213 pKa = 3.45MMDD216 pKa = 4.0YY217 pKa = 10.48FISAIPNTWSGDD229 pKa = 3.76SPVALHH235 pKa = 6.01VALPEE240 pKa = 4.25GQVILNDD247 pKa = 3.53INSGADD253 pKa = 2.93IRR255 pKa = 11.84VVHH258 pKa = 6.6AVADD262 pKa = 4.18APAVDD267 pKa = 3.87VFLDD271 pKa = 3.83EE272 pKa = 4.53ATTPAIDD279 pKa = 3.29MLEE282 pKa = 4.01FGKK285 pKa = 10.48IAGYY289 pKa = 10.86VNVAEE294 pKa = 4.74GQHH297 pKa = 5.2TVTVAADD304 pKa = 3.5ADD306 pKa = 3.88NSVVVIDD313 pKa = 4.08KK314 pKa = 11.26APVEE318 pKa = 4.11LMLGMSYY325 pKa = 10.65SALAIGSLSDD335 pKa = 4.09GMIEE339 pKa = 4.16PLVLTEE345 pKa = 3.59KK346 pKa = 9.4TRR348 pKa = 11.84RR349 pKa = 11.84VATEE353 pKa = 3.53AKK355 pKa = 9.53LTVTHH360 pKa = 7.03AAYY363 pKa = 10.2SAPEE367 pKa = 3.54VDD369 pKa = 4.29IYY371 pKa = 10.87LTATADD377 pKa = 3.2ISEE380 pKa = 4.45ATPALEE386 pKa = 4.58DD387 pKa = 3.73VPFKK391 pKa = 10.98ASSGSLSVAPGTYY404 pKa = 9.37TISVTVANTKK414 pKa = 10.27DD415 pKa = 3.54VAIGPLEE422 pKa = 4.22VTLEE426 pKa = 3.92AAGVYY431 pKa = 9.96GVAAVDD437 pKa = 3.77NVGGGAPFNVILLDD451 pKa = 4.29DD452 pKa = 4.1FTVMM456 pKa = 5.18
MM1 pKa = 7.34KK2 pKa = 10.22RR3 pKa = 11.84IYY5 pKa = 10.35PFALCISLVGLTACSDD21 pKa = 3.76DD22 pKa = 5.7DD23 pKa = 4.38DD24 pKa = 5.99DD25 pKa = 6.2KK26 pKa = 11.93VVTPDD31 pKa = 3.43TPEE34 pKa = 3.59VEE36 pKa = 4.4YY37 pKa = 11.07SHH39 pKa = 7.04VRR41 pKa = 11.84VIHH44 pKa = 6.39AGSDD48 pKa = 3.19APMVNVMANGAALLSDD64 pKa = 3.43VDD66 pKa = 4.02YY67 pKa = 11.77AMSSGLLEE75 pKa = 4.35VTSATYY81 pKa = 10.45DD82 pKa = 3.25IDD84 pKa = 4.1VDD86 pKa = 3.92ALLADD91 pKa = 4.21GTTLTVLEE99 pKa = 4.87ANLATEE105 pKa = 4.66ANTEE109 pKa = 4.11YY110 pKa = 10.31TAVALGTVADD120 pKa = 4.37EE121 pKa = 4.63SLMLKK126 pKa = 10.55LIANPTAEE134 pKa = 4.0IAAGYY139 pKa = 10.84ARR141 pKa = 11.84VQVLHH146 pKa = 6.2ATPSVGLVDD155 pKa = 4.21VYY157 pKa = 10.1VTAPGDD163 pKa = 4.44DD164 pKa = 3.39ISAIAPTLSANYY176 pKa = 8.8MDD178 pKa = 5.48NSTQLEE184 pKa = 4.71VPVGDD189 pKa = 3.54YY190 pKa = 10.46QIRR193 pKa = 11.84ITGSNSKK200 pKa = 10.33EE201 pKa = 3.88VVFDD205 pKa = 3.61SGTVALGDD213 pKa = 3.45MMDD216 pKa = 4.0YY217 pKa = 10.48FISAIPNTWSGDD229 pKa = 3.76SPVALHH235 pKa = 6.01VALPEE240 pKa = 4.25GQVILNDD247 pKa = 3.53INSGADD253 pKa = 2.93IRR255 pKa = 11.84VVHH258 pKa = 6.6AVADD262 pKa = 4.18APAVDD267 pKa = 3.87VFLDD271 pKa = 3.83EE272 pKa = 4.53ATTPAIDD279 pKa = 3.29MLEE282 pKa = 4.01FGKK285 pKa = 10.48IAGYY289 pKa = 10.86VNVAEE294 pKa = 4.74GQHH297 pKa = 5.2TVTVAADD304 pKa = 3.5ADD306 pKa = 3.88NSVVVIDD313 pKa = 4.08KK314 pKa = 11.26APVEE318 pKa = 4.11LMLGMSYY325 pKa = 10.65SALAIGSLSDD335 pKa = 4.09GMIEE339 pKa = 4.16PLVLTEE345 pKa = 3.59KK346 pKa = 9.4TRR348 pKa = 11.84RR349 pKa = 11.84VATEE353 pKa = 3.53AKK355 pKa = 9.53LTVTHH360 pKa = 7.03AAYY363 pKa = 10.2SAPEE367 pKa = 3.54VDD369 pKa = 4.29IYY371 pKa = 10.87LTATADD377 pKa = 3.2ISEE380 pKa = 4.45ATPALEE386 pKa = 4.58DD387 pKa = 3.73VPFKK391 pKa = 10.98ASSGSLSVAPGTYY404 pKa = 9.37TISVTVANTKK414 pKa = 10.27DD415 pKa = 3.54VAIGPLEE422 pKa = 4.22VTLEE426 pKa = 3.92AAGVYY431 pKa = 9.96GVAAVDD437 pKa = 3.77NVGGGAPFNVILLDD451 pKa = 4.29DD452 pKa = 4.1FTVMM456 pKa = 5.18
Molecular weight: 47.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G9QNB1|A0A6G9QNB1_9GAMM YacL family protein OS=Shewanella aestuarii OX=1028752 GN=HBH39_14565 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MATAGGRR29 pKa = 11.84KK30 pKa = 8.78VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.39GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MATAGGRR29 pKa = 11.84KK30 pKa = 8.78VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.39GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1213747 |
33 |
4547 |
336.7 |
37.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.806 ± 0.044 | 1.085 ± 0.019 |
5.706 ± 0.034 | 5.707 ± 0.037 |
4.129 ± 0.027 | 6.541 ± 0.039 |
2.267 ± 0.022 | 6.712 ± 0.037 |
5.423 ± 0.043 | 10.265 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.603 ± 0.021 | 4.715 ± 0.042 |
3.8 ± 0.023 | 5.065 ± 0.041 |
4.117 ± 0.037 | 6.687 ± 0.045 |
5.433 ± 0.047 | 6.745 ± 0.032 |
1.135 ± 0.015 | 3.06 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
