
Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Legionellales; Legionellaceae; Legionella; Legionella pneumophila; Legionella pneumophila subsp. pneumophila
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
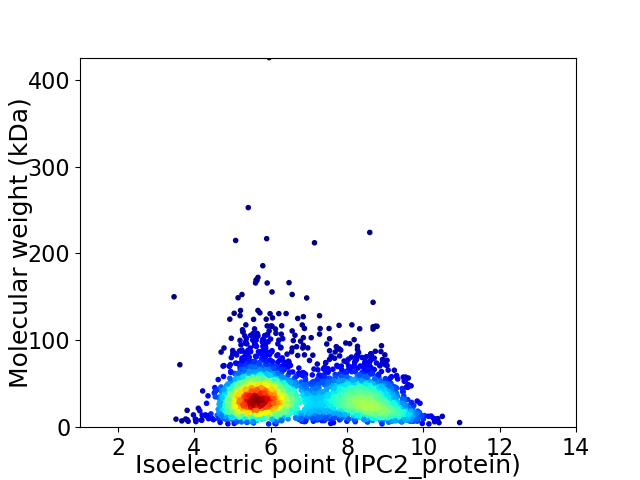
Virtual 2D-PAGE plot for 2930 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5ZXV6|Q5ZXV6_LEGPH Expressed protein OS=Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513) OX=272624 GN=lpg0625 PE=4 SV=1
MM1 pKa = 7.32LAEE4 pKa = 4.1SVIGIVRR11 pKa = 11.84AVNGLLEE18 pKa = 4.23KK19 pKa = 11.24VNVQGQASLVKK30 pKa = 10.27SGARR34 pKa = 11.84LEE36 pKa = 4.47EE37 pKa = 4.9GDD39 pKa = 4.43VLTLLSGEE47 pKa = 4.59AYY49 pKa = 9.54IQFIHH54 pKa = 6.82GFPEE58 pKa = 3.96ALALEE63 pKa = 4.75KK64 pKa = 10.12PVKK67 pKa = 10.35LDD69 pKa = 3.42GVSPTLQYY77 pKa = 10.98GAEE80 pKa = 4.01DD81 pKa = 3.65LKK83 pKa = 11.38EE84 pKa = 4.03QMVQEE89 pKa = 5.21AIAKK93 pKa = 10.16GIDD96 pKa = 3.28PSVILDD102 pKa = 3.73VLGSAAAGAVAVGSGGDD119 pKa = 3.52AFIIDD124 pKa = 3.81PLFGFGHH131 pKa = 5.4VTAGYY136 pKa = 6.21PTGPISFAYY145 pKa = 9.89EE146 pKa = 3.85ADD148 pKa = 3.73TQQLFWFVPEE158 pKa = 3.91EE159 pKa = 3.86TGAIAEE165 pKa = 4.49SEE167 pKa = 4.25LTTEE171 pKa = 5.02PEE173 pKa = 4.13FTPQIPQFTTNQAVLTVFEE192 pKa = 5.06DD193 pKa = 3.97ALPSGIPDD201 pKa = 3.25SAGQARR207 pKa = 11.84TASSSLSTLLTSSPDD222 pKa = 3.34VAASFAFNTNLSVLPTLKK240 pKa = 10.65SGGIDD245 pKa = 3.45LDD247 pKa = 4.45YY248 pKa = 11.36NLSPDD253 pKa = 3.6KK254 pKa = 10.39RR255 pKa = 11.84TLTASIPGGGDD266 pKa = 3.04VMQFEE271 pKa = 4.77LTADD275 pKa = 3.41GRR277 pKa = 11.84LTQTLMDD284 pKa = 5.44SIDD287 pKa = 3.83HH288 pKa = 6.72PTTDD292 pKa = 3.53SDD294 pKa = 3.58DD295 pKa = 4.49GEE297 pKa = 4.35WMRR300 pKa = 11.84LDD302 pKa = 4.61LSPLIYY308 pKa = 10.12VTFTRR313 pKa = 11.84TSDD316 pKa = 3.52GTVLEE321 pKa = 4.53SRR323 pKa = 11.84TLPANAVVAGIQDD336 pKa = 3.76DD337 pKa = 4.34VPIARR342 pKa = 11.84AQLTNNEE349 pKa = 4.06ILLDD353 pKa = 3.6EE354 pKa = 4.4TVGVKK359 pKa = 10.48VGDD362 pKa = 3.68ADD364 pKa = 4.66AANDD368 pKa = 4.25DD369 pKa = 4.42FNPTITTDD377 pKa = 3.65PFNNTYY383 pKa = 10.4GIPIGLVQNANLLDD397 pKa = 4.11NGTSEE402 pKa = 4.66MGGDD406 pKa = 3.71YY407 pKa = 11.03KK408 pKa = 11.05NATMTHH414 pKa = 6.62LLKK417 pKa = 9.93ITDD420 pKa = 3.99AVSGLQTTDD429 pKa = 2.73GTSINLFLEE438 pKa = 4.43SNGDD442 pKa = 3.06ITGRR446 pKa = 11.84AGDD449 pKa = 3.44IGEE452 pKa = 4.26PAVFAIRR459 pKa = 11.84MNPNTGSITVAQYY472 pKa = 11.6GSIKK476 pKa = 10.54QFDD479 pKa = 4.28TNSHH483 pKa = 6.79DD484 pKa = 3.79EE485 pKa = 4.28AVDD488 pKa = 3.48LTGRR492 pKa = 11.84ISAVVTAKK500 pKa = 10.92DD501 pKa = 3.15SDD503 pKa = 3.89GDD505 pKa = 3.76VSNVEE510 pKa = 3.87IPIGQLIIFADD521 pKa = 4.47DD522 pKa = 4.35GPSVTMAVSDD532 pKa = 4.01NNAITLNTQDD542 pKa = 4.44ADD544 pKa = 4.34TIGAASDD551 pKa = 3.51SDD553 pKa = 4.01SASFAAAFAVTPNYY567 pKa = 10.22GADD570 pKa = 3.64GAGTTVTTYY579 pKa = 11.08ALSVSAQGVDD589 pKa = 3.71SGLDD593 pKa = 3.28NNGNNIYY600 pKa = 9.96LYY602 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD629 pKa = 3.69VNSSSGVVTLTQHH642 pKa = 5.98QEE644 pKa = 3.71VDD646 pKa = 3.11HH647 pKa = 6.39GLPGASSNYY656 pKa = 9.21AAQEE660 pKa = 4.5AILNTGLVFLNATAVTTDD678 pKa = 3.21GDD680 pKa = 4.1GDD682 pKa = 3.96TATASASLDD691 pKa = 3.2LGGNVKK697 pKa = 10.18FDD699 pKa = 4.29DD700 pKa = 5.2DD701 pKa = 4.5GPSVTMAVSDD711 pKa = 4.01NNAITLNTQDD721 pKa = 4.44ADD723 pKa = 4.34TIGAASDD730 pKa = 3.51SDD732 pKa = 4.01SASFAAAFAVTPNYY746 pKa = 10.22GADD749 pKa = 3.64GAGTTVTTYY758 pKa = 11.08ALSVSAQGVDD768 pKa = 3.71SGLDD772 pKa = 3.28NNGNNIYY779 pKa = 9.96LYY781 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD808 pKa = 3.69VNSSSGVVTLTQHH821 pKa = 5.98QEE823 pKa = 3.71VDD825 pKa = 3.11HH826 pKa = 6.39GLPGASSNYY835 pKa = 9.21AAQEE839 pKa = 4.5AILNTGLVFLNATAVTTDD857 pKa = 3.21GDD859 pKa = 4.1GDD861 pKa = 3.96TATASASLDD870 pKa = 3.2LGGNVKK876 pKa = 10.18FDD878 pKa = 4.29DD879 pKa = 5.2DD880 pKa = 4.5GPSVTMAVSDD890 pKa = 4.01NNAITLNTQDD900 pKa = 4.44ADD902 pKa = 4.34TIGAASDD909 pKa = 3.51SDD911 pKa = 4.01SASFAAAFAVTPNYY925 pKa = 10.22GADD928 pKa = 3.64GAGTTVTTYY937 pKa = 11.08ALSVSAQGVDD947 pKa = 3.71SGLDD951 pKa = 3.28NNGNNIYY958 pKa = 9.96LYY960 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD987 pKa = 3.69VNSSSGVVTLTQHH1000 pKa = 5.98QEE1002 pKa = 3.71VDD1004 pKa = 3.11HH1005 pKa = 6.39GLPGASSNYY1014 pKa = 9.21AAQEE1018 pKa = 4.5AILNTGLVFLNATAVTTDD1036 pKa = 3.21GDD1038 pKa = 4.1GDD1040 pKa = 3.96TATASASLDD1049 pKa = 3.2LGGNVKK1055 pKa = 10.18FDD1057 pKa = 4.29DD1058 pKa = 5.2DD1059 pKa = 4.5GPSVTMAVSDD1069 pKa = 4.01NNAITLNTQDD1079 pKa = 4.06AEE1081 pKa = 4.71TIGVLSDD1088 pKa = 3.14TDD1090 pKa = 3.87SASFAAAFAVTPNYY1104 pKa = 10.22GADD1107 pKa = 3.64GAGTTVTTYY1116 pKa = 11.08ALSVSAQGVDD1126 pKa = 3.71SGLDD1130 pKa = 3.28NNGNNIYY1137 pKa = 9.96LYY1139 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD1166 pKa = 3.69VNSSSGVVTLTQHH1179 pKa = 5.98QEE1181 pKa = 3.71VDD1183 pKa = 3.11HH1184 pKa = 6.39GLPGASSNYY1193 pKa = 9.21AAQEE1197 pKa = 4.5AILNTGLVFLNATAVTTDD1215 pKa = 3.21GDD1217 pKa = 4.1GDD1219 pKa = 3.96TATASASLDD1228 pKa = 3.2LGGNVKK1234 pKa = 10.18FDD1236 pKa = 4.29DD1237 pKa = 5.2DD1238 pKa = 4.5GPSVTMAVSDD1248 pKa = 4.01NNAITLNTQDD1258 pKa = 4.44ADD1260 pKa = 4.34TIGAASDD1267 pKa = 3.51SDD1269 pKa = 4.01SASFAAAFAVTPNYY1283 pKa = 10.22GADD1286 pKa = 3.64GAGTTVTTYY1295 pKa = 11.08ALSVSAQGVDD1305 pKa = 3.71SGLDD1309 pKa = 3.28NNGNNIYY1316 pKa = 9.96LYY1318 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD1345 pKa = 3.69VNSSSGVVTLTQHH1358 pKa = 5.98QEE1360 pKa = 3.71VDD1362 pKa = 3.11HH1363 pKa = 6.39GLPGASSNYY1372 pKa = 9.21AAQEE1376 pKa = 4.5AILNTGLVFLNATAVTTDD1394 pKa = 3.21GDD1396 pKa = 4.1GDD1398 pKa = 3.96TATASASLDD1407 pKa = 3.2LGGNVKK1413 pKa = 10.18FDD1415 pKa = 4.29DD1416 pKa = 5.2DD1417 pKa = 4.5GPSVTMAVSDD1427 pKa = 4.01NNAITLNTQDD1437 pKa = 4.44ADD1439 pKa = 4.34TIGAASDD1446 pKa = 3.51SDD1448 pKa = 4.01SASFAAAFAVTPNYY1462 pKa = 10.22GADD1465 pKa = 3.64GAGTTVTTYY1474 pKa = 10.93ALSVSAPRR1482 pKa = 11.84GGFRR1486 pKa = 11.84PP1487 pKa = 3.7
MM1 pKa = 7.32LAEE4 pKa = 4.1SVIGIVRR11 pKa = 11.84AVNGLLEE18 pKa = 4.23KK19 pKa = 11.24VNVQGQASLVKK30 pKa = 10.27SGARR34 pKa = 11.84LEE36 pKa = 4.47EE37 pKa = 4.9GDD39 pKa = 4.43VLTLLSGEE47 pKa = 4.59AYY49 pKa = 9.54IQFIHH54 pKa = 6.82GFPEE58 pKa = 3.96ALALEE63 pKa = 4.75KK64 pKa = 10.12PVKK67 pKa = 10.35LDD69 pKa = 3.42GVSPTLQYY77 pKa = 10.98GAEE80 pKa = 4.01DD81 pKa = 3.65LKK83 pKa = 11.38EE84 pKa = 4.03QMVQEE89 pKa = 5.21AIAKK93 pKa = 10.16GIDD96 pKa = 3.28PSVILDD102 pKa = 3.73VLGSAAAGAVAVGSGGDD119 pKa = 3.52AFIIDD124 pKa = 3.81PLFGFGHH131 pKa = 5.4VTAGYY136 pKa = 6.21PTGPISFAYY145 pKa = 9.89EE146 pKa = 3.85ADD148 pKa = 3.73TQQLFWFVPEE158 pKa = 3.91EE159 pKa = 3.86TGAIAEE165 pKa = 4.49SEE167 pKa = 4.25LTTEE171 pKa = 5.02PEE173 pKa = 4.13FTPQIPQFTTNQAVLTVFEE192 pKa = 5.06DD193 pKa = 3.97ALPSGIPDD201 pKa = 3.25SAGQARR207 pKa = 11.84TASSSLSTLLTSSPDD222 pKa = 3.34VAASFAFNTNLSVLPTLKK240 pKa = 10.65SGGIDD245 pKa = 3.45LDD247 pKa = 4.45YY248 pKa = 11.36NLSPDD253 pKa = 3.6KK254 pKa = 10.39RR255 pKa = 11.84TLTASIPGGGDD266 pKa = 3.04VMQFEE271 pKa = 4.77LTADD275 pKa = 3.41GRR277 pKa = 11.84LTQTLMDD284 pKa = 5.44SIDD287 pKa = 3.83HH288 pKa = 6.72PTTDD292 pKa = 3.53SDD294 pKa = 3.58DD295 pKa = 4.49GEE297 pKa = 4.35WMRR300 pKa = 11.84LDD302 pKa = 4.61LSPLIYY308 pKa = 10.12VTFTRR313 pKa = 11.84TSDD316 pKa = 3.52GTVLEE321 pKa = 4.53SRR323 pKa = 11.84TLPANAVVAGIQDD336 pKa = 3.76DD337 pKa = 4.34VPIARR342 pKa = 11.84AQLTNNEE349 pKa = 4.06ILLDD353 pKa = 3.6EE354 pKa = 4.4TVGVKK359 pKa = 10.48VGDD362 pKa = 3.68ADD364 pKa = 4.66AANDD368 pKa = 4.25DD369 pKa = 4.42FNPTITTDD377 pKa = 3.65PFNNTYY383 pKa = 10.4GIPIGLVQNANLLDD397 pKa = 4.11NGTSEE402 pKa = 4.66MGGDD406 pKa = 3.71YY407 pKa = 11.03KK408 pKa = 11.05NATMTHH414 pKa = 6.62LLKK417 pKa = 9.93ITDD420 pKa = 3.99AVSGLQTTDD429 pKa = 2.73GTSINLFLEE438 pKa = 4.43SNGDD442 pKa = 3.06ITGRR446 pKa = 11.84AGDD449 pKa = 3.44IGEE452 pKa = 4.26PAVFAIRR459 pKa = 11.84MNPNTGSITVAQYY472 pKa = 11.6GSIKK476 pKa = 10.54QFDD479 pKa = 4.28TNSHH483 pKa = 6.79DD484 pKa = 3.79EE485 pKa = 4.28AVDD488 pKa = 3.48LTGRR492 pKa = 11.84ISAVVTAKK500 pKa = 10.92DD501 pKa = 3.15SDD503 pKa = 3.89GDD505 pKa = 3.76VSNVEE510 pKa = 3.87IPIGQLIIFADD521 pKa = 4.47DD522 pKa = 4.35GPSVTMAVSDD532 pKa = 4.01NNAITLNTQDD542 pKa = 4.44ADD544 pKa = 4.34TIGAASDD551 pKa = 3.51SDD553 pKa = 4.01SASFAAAFAVTPNYY567 pKa = 10.22GADD570 pKa = 3.64GAGTTVTTYY579 pKa = 11.08ALSVSAQGVDD589 pKa = 3.71SGLDD593 pKa = 3.28NNGNNIYY600 pKa = 9.96LYY602 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD629 pKa = 3.69VNSSSGVVTLTQHH642 pKa = 5.98QEE644 pKa = 3.71VDD646 pKa = 3.11HH647 pKa = 6.39GLPGASSNYY656 pKa = 9.21AAQEE660 pKa = 4.5AILNTGLVFLNATAVTTDD678 pKa = 3.21GDD680 pKa = 4.1GDD682 pKa = 3.96TATASASLDD691 pKa = 3.2LGGNVKK697 pKa = 10.18FDD699 pKa = 4.29DD700 pKa = 5.2DD701 pKa = 4.5GPSVTMAVSDD711 pKa = 4.01NNAITLNTQDD721 pKa = 4.44ADD723 pKa = 4.34TIGAASDD730 pKa = 3.51SDD732 pKa = 4.01SASFAAAFAVTPNYY746 pKa = 10.22GADD749 pKa = 3.64GAGTTVTTYY758 pKa = 11.08ALSVSAQGVDD768 pKa = 3.71SGLDD772 pKa = 3.28NNGNNIYY779 pKa = 9.96LYY781 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD808 pKa = 3.69VNSSSGVVTLTQHH821 pKa = 5.98QEE823 pKa = 3.71VDD825 pKa = 3.11HH826 pKa = 6.39GLPGASSNYY835 pKa = 9.21AAQEE839 pKa = 4.5AILNTGLVFLNATAVTTDD857 pKa = 3.21GDD859 pKa = 4.1GDD861 pKa = 3.96TATASASLDD870 pKa = 3.2LGGNVKK876 pKa = 10.18FDD878 pKa = 4.29DD879 pKa = 5.2DD880 pKa = 4.5GPSVTMAVSDD890 pKa = 4.01NNAITLNTQDD900 pKa = 4.44ADD902 pKa = 4.34TIGAASDD909 pKa = 3.51SDD911 pKa = 4.01SASFAAAFAVTPNYY925 pKa = 10.22GADD928 pKa = 3.64GAGTTVTTYY937 pKa = 11.08ALSVSAQGVDD947 pKa = 3.71SGLDD951 pKa = 3.28NNGNNIYY958 pKa = 9.96LYY960 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD987 pKa = 3.69VNSSSGVVTLTQHH1000 pKa = 5.98QEE1002 pKa = 3.71VDD1004 pKa = 3.11HH1005 pKa = 6.39GLPGASSNYY1014 pKa = 9.21AAQEE1018 pKa = 4.5AILNTGLVFLNATAVTTDD1036 pKa = 3.21GDD1038 pKa = 4.1GDD1040 pKa = 3.96TATASASLDD1049 pKa = 3.2LGGNVKK1055 pKa = 10.18FDD1057 pKa = 4.29DD1058 pKa = 5.2DD1059 pKa = 4.5GPSVTMAVSDD1069 pKa = 4.01NNAITLNTQDD1079 pKa = 4.06AEE1081 pKa = 4.71TIGVLSDD1088 pKa = 3.14TDD1090 pKa = 3.87SASFAAAFAVTPNYY1104 pKa = 10.22GADD1107 pKa = 3.64GAGTTVTTYY1116 pKa = 11.08ALSVSAQGVDD1126 pKa = 3.71SGLDD1130 pKa = 3.28NNGNNIYY1137 pKa = 9.96LYY1139 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD1166 pKa = 3.69VNSSSGVVTLTQHH1179 pKa = 5.98QEE1181 pKa = 3.71VDD1183 pKa = 3.11HH1184 pKa = 6.39GLPGASSNYY1193 pKa = 9.21AAQEE1197 pKa = 4.5AILNTGLVFLNATAVTTDD1215 pKa = 3.21GDD1217 pKa = 4.1GDD1219 pKa = 3.96TATASASLDD1228 pKa = 3.2LGGNVKK1234 pKa = 10.18FDD1236 pKa = 4.29DD1237 pKa = 5.2DD1238 pKa = 4.5GPSVTMAVSDD1248 pKa = 4.01NNAITLNTQDD1258 pKa = 4.44ADD1260 pKa = 4.34TIGAASDD1267 pKa = 3.51SDD1269 pKa = 4.01SASFAAAFAVTPNYY1283 pKa = 10.22GADD1286 pKa = 3.64GAGTTVTTYY1295 pKa = 11.08ALSVSAQGVDD1305 pKa = 3.71SGLDD1309 pKa = 3.28NNGNNIYY1316 pKa = 9.96LYY1318 pKa = 10.07NIAGSVVGSTSATQAGITTGNTIFSLDD1345 pKa = 3.69VNSSSGVVTLTQHH1358 pKa = 5.98QEE1360 pKa = 3.71VDD1362 pKa = 3.11HH1363 pKa = 6.39GLPGASSNYY1372 pKa = 9.21AAQEE1376 pKa = 4.5AILNTGLVFLNATAVTTDD1394 pKa = 3.21GDD1396 pKa = 4.1GDD1398 pKa = 3.96TATASASLDD1407 pKa = 3.2LGGNVKK1413 pKa = 10.18FDD1415 pKa = 4.29DD1416 pKa = 5.2DD1417 pKa = 4.5GPSVTMAVSDD1427 pKa = 4.01NNAITLNTQDD1437 pKa = 4.44ADD1439 pKa = 4.34TIGAASDD1446 pKa = 3.51SDD1448 pKa = 4.01SASFAAAFAVTPNYY1462 pKa = 10.22GADD1465 pKa = 3.64GAGTTVTTYY1474 pKa = 10.93ALSVSAPRR1482 pKa = 11.84GGFRR1486 pKa = 11.84PP1487 pKa = 3.7
Molecular weight: 150.1 kDa
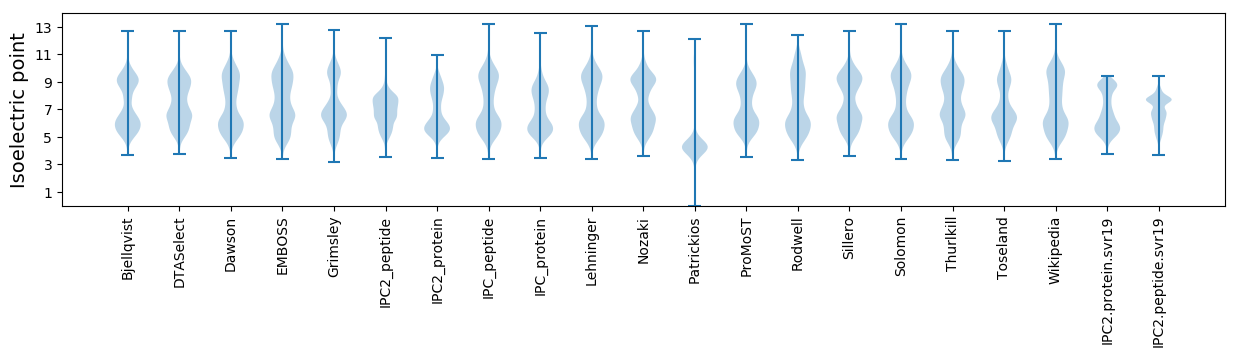
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5ZR83|Q5ZR83_LEGPH Uncharacterized protein OS=Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513) OX=272624 GN=lpg3000 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.18RR12 pKa = 11.84KK13 pKa = 8.9RR14 pKa = 11.84DD15 pKa = 3.28HH16 pKa = 6.64GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTRR25 pKa = 11.84AGRR28 pKa = 11.84LVIKK32 pKa = 10.41RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.18RR12 pKa = 11.84KK13 pKa = 8.9RR14 pKa = 11.84DD15 pKa = 3.28HH16 pKa = 6.64GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTRR25 pKa = 11.84AGRR28 pKa = 11.84LVIKK32 pKa = 10.41RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
997422 |
34 |
3780 |
340.4 |
38.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.198 ± 0.048 | 1.21 ± 0.018 |
4.821 ± 0.033 | 6.031 ± 0.05 |
4.501 ± 0.03 | 6.035 ± 0.044 |
2.464 ± 0.021 | 7.628 ± 0.042 |
6.507 ± 0.044 | 10.957 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.465 ± 0.019 | 5.01 ± 0.032 |
4.062 ± 0.028 | 4.536 ± 0.037 |
4.161 ± 0.033 | 6.68 ± 0.034 |
5.137 ± 0.029 | 5.88 ± 0.039 |
1.151 ± 0.017 | 3.567 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
