
Felis catus papillomavirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Taupapillomavirus; Taupapillomavirus 3
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
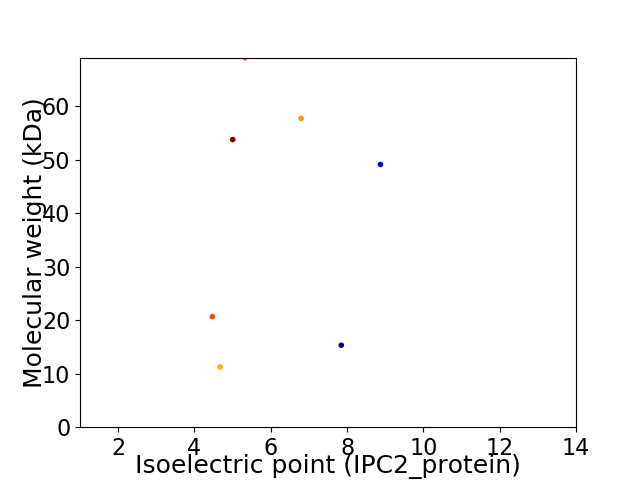
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T2D2T1|T2D2T1_9PAPI Major capsid protein L1 OS=Felis catus papillomavirus 4 OX=1398507 GN=L1 PE=3 SV=1
MM1 pKa = 7.51IGKK4 pKa = 8.89EE5 pKa = 3.94PQIKK9 pKa = 10.27DD10 pKa = 3.04IEE12 pKa = 5.04LDD14 pKa = 3.47LDD16 pKa = 4.1EE17 pKa = 5.82LVLPANLLASEE28 pKa = 5.1EE29 pKa = 3.97ILEE32 pKa = 4.37AEE34 pKa = 4.31EE35 pKa = 4.28EE36 pKa = 4.4EE37 pKa = 4.52EE38 pKa = 4.67PEE40 pKa = 4.28SNPYY44 pKa = 10.3RR45 pKa = 11.84IVTCCVLCHH54 pKa = 5.24STLRR58 pKa = 11.84LVVSATDD65 pKa = 3.44AQIRR69 pKa = 11.84AQQDD73 pKa = 3.28LFLAGLGIICPVCYY87 pKa = 10.12RR88 pKa = 11.84NNRR91 pKa = 11.84NHH93 pKa = 7.32GGQQGKK99 pKa = 9.6RR100 pKa = 11.84PP101 pKa = 3.63
MM1 pKa = 7.51IGKK4 pKa = 8.89EE5 pKa = 3.94PQIKK9 pKa = 10.27DD10 pKa = 3.04IEE12 pKa = 5.04LDD14 pKa = 3.47LDD16 pKa = 4.1EE17 pKa = 5.82LVLPANLLASEE28 pKa = 5.1EE29 pKa = 3.97ILEE32 pKa = 4.37AEE34 pKa = 4.31EE35 pKa = 4.28EE36 pKa = 4.4EE37 pKa = 4.52EE38 pKa = 4.67PEE40 pKa = 4.28SNPYY44 pKa = 10.3RR45 pKa = 11.84IVTCCVLCHH54 pKa = 5.24STLRR58 pKa = 11.84LVVSATDD65 pKa = 3.44AQIRR69 pKa = 11.84AQQDD73 pKa = 3.28LFLAGLGIICPVCYY87 pKa = 10.12RR88 pKa = 11.84NNRR91 pKa = 11.84NHH93 pKa = 7.32GGQQGKK99 pKa = 9.6RR100 pKa = 11.84PP101 pKa = 3.63
Molecular weight: 11.24 kDa
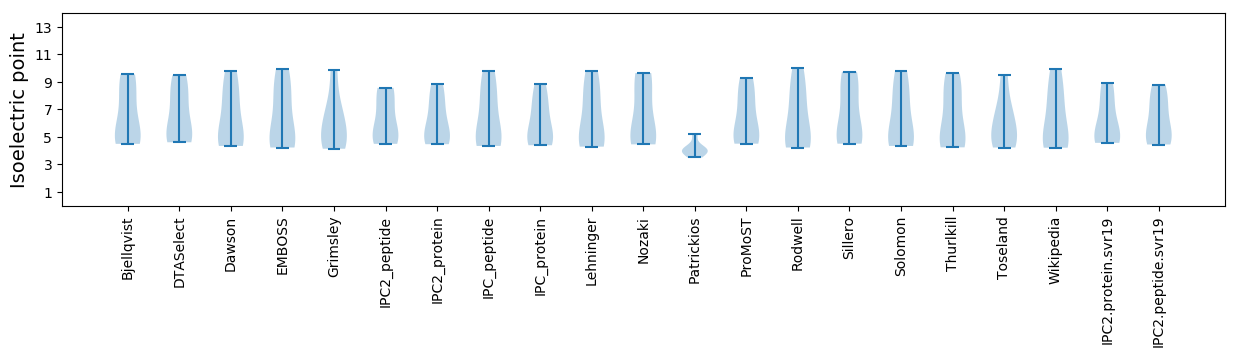
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T2D3L1|T2D3L1_9PAPI Putative E4 protein OS=Felis catus papillomavirus 4 OX=1398507 GN=E4 PE=4 SV=1
MM1 pKa = 7.71EE2 pKa = 4.95NLSTRR7 pKa = 11.84FDD9 pKa = 3.69TLQEE13 pKa = 3.93VLLAHH18 pKa = 5.91YY19 pKa = 9.33EE20 pKa = 4.03KK21 pKa = 10.48DD22 pKa = 3.41SKK24 pKa = 11.18KK25 pKa = 9.39ITDD28 pKa = 3.82HH29 pKa = 5.96VSFWEE34 pKa = 3.78LLRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.45SVMLHH44 pKa = 5.78YY45 pKa = 10.61ARR47 pKa = 11.84QQGISSLGFFQVPSLQVSEE66 pKa = 5.19AKK68 pKa = 10.01AKK70 pKa = 9.73KK71 pKa = 9.98AIMMSLMLQQLAKK84 pKa = 9.98TPFGQEE90 pKa = 3.64PWSMTEE96 pKa = 3.71TSLEE100 pKa = 4.02MLEE103 pKa = 4.84APPKK107 pKa = 10.63GKK109 pKa = 9.91FKK111 pKa = 10.76KK112 pKa = 10.42GPRR115 pKa = 11.84TVEE118 pKa = 3.46VWFDD122 pKa = 3.81NNPDD126 pKa = 3.05NSFPYY131 pKa = 10.15TSWTCIYY138 pKa = 10.23IQDD141 pKa = 5.57AEE143 pKa = 4.48DD144 pKa = 3.17TWHH147 pKa = 6.58KK148 pKa = 11.3VEE150 pKa = 4.93GLVDD154 pKa = 3.9YY155 pKa = 10.78EE156 pKa = 3.79GLYY159 pKa = 10.74YY160 pKa = 10.34VDD162 pKa = 4.82CDD164 pKa = 4.49GEE166 pKa = 3.97VHH168 pKa = 6.46YY169 pKa = 10.21YY170 pKa = 10.54VKK172 pKa = 10.5FAADD176 pKa = 3.43ALQYY180 pKa = 9.54ATTGMWRR187 pKa = 11.84VNYY190 pKa = 9.98KK191 pKa = 9.96NQTISASVSSSSSEE205 pKa = 3.78DD206 pKa = 3.25QQRR209 pKa = 11.84QGQQQQQQQQQPSTTTSVEE228 pKa = 3.82WPQRR232 pKa = 11.84PSSSGLQHH240 pKa = 6.83EE241 pKa = 5.16SPFTPRR247 pKa = 11.84GHH249 pKa = 6.69GRR251 pKa = 11.84GCVRR255 pKa = 11.84GRR257 pKa = 11.84RR258 pKa = 11.84SSGSSPHH265 pKa = 5.4QRR267 pKa = 11.84GDD269 pKa = 2.76RR270 pKa = 11.84GGRR273 pKa = 11.84GRR275 pKa = 11.84SQSPSTPTTPGALPPDD291 pKa = 4.07LSFAGGDD298 pKa = 3.22RR299 pKa = 11.84SGGGGGGGGGGRR311 pKa = 11.84QRR313 pKa = 11.84GRR315 pKa = 11.84RR316 pKa = 11.84GSRR319 pKa = 11.84GPPRR323 pKa = 11.84LSPYY327 pKa = 9.95IPLGQVEE334 pKa = 4.43QGPGRR339 pKa = 11.84PEE341 pKa = 3.67GAALQRR347 pKa = 11.84PGRR350 pKa = 11.84PEE352 pKa = 3.59KK353 pKa = 10.75NPRR356 pKa = 11.84LSSTTPVVVLKK367 pKa = 11.19GPGNALKK374 pKa = 10.35CWRR377 pKa = 11.84LRR379 pKa = 11.84AKK381 pKa = 10.1AKK383 pKa = 10.45HH384 pKa = 5.65GALFCAISTAFSWVEE399 pKa = 3.66KK400 pKa = 10.25SSSSRR405 pKa = 11.84IGRR408 pKa = 11.84HH409 pKa = 5.7RR410 pKa = 11.84ILVGFINEE418 pKa = 4.0EE419 pKa = 3.71QRR421 pKa = 11.84EE422 pKa = 4.23DD423 pKa = 3.46FLRR426 pKa = 11.84TVRR429 pKa = 11.84LPRR432 pKa = 11.84GVEE435 pKa = 4.12CVPGGLDD442 pKa = 3.5SLL444 pKa = 4.5
MM1 pKa = 7.71EE2 pKa = 4.95NLSTRR7 pKa = 11.84FDD9 pKa = 3.69TLQEE13 pKa = 3.93VLLAHH18 pKa = 5.91YY19 pKa = 9.33EE20 pKa = 4.03KK21 pKa = 10.48DD22 pKa = 3.41SKK24 pKa = 11.18KK25 pKa = 9.39ITDD28 pKa = 3.82HH29 pKa = 5.96VSFWEE34 pKa = 3.78LLRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.45SVMLHH44 pKa = 5.78YY45 pKa = 10.61ARR47 pKa = 11.84QQGISSLGFFQVPSLQVSEE66 pKa = 5.19AKK68 pKa = 10.01AKK70 pKa = 9.73KK71 pKa = 9.98AIMMSLMLQQLAKK84 pKa = 9.98TPFGQEE90 pKa = 3.64PWSMTEE96 pKa = 3.71TSLEE100 pKa = 4.02MLEE103 pKa = 4.84APPKK107 pKa = 10.63GKK109 pKa = 9.91FKK111 pKa = 10.76KK112 pKa = 10.42GPRR115 pKa = 11.84TVEE118 pKa = 3.46VWFDD122 pKa = 3.81NNPDD126 pKa = 3.05NSFPYY131 pKa = 10.15TSWTCIYY138 pKa = 10.23IQDD141 pKa = 5.57AEE143 pKa = 4.48DD144 pKa = 3.17TWHH147 pKa = 6.58KK148 pKa = 11.3VEE150 pKa = 4.93GLVDD154 pKa = 3.9YY155 pKa = 10.78EE156 pKa = 3.79GLYY159 pKa = 10.74YY160 pKa = 10.34VDD162 pKa = 4.82CDD164 pKa = 4.49GEE166 pKa = 3.97VHH168 pKa = 6.46YY169 pKa = 10.21YY170 pKa = 10.54VKK172 pKa = 10.5FAADD176 pKa = 3.43ALQYY180 pKa = 9.54ATTGMWRR187 pKa = 11.84VNYY190 pKa = 9.98KK191 pKa = 9.96NQTISASVSSSSSEE205 pKa = 3.78DD206 pKa = 3.25QQRR209 pKa = 11.84QGQQQQQQQQQPSTTTSVEE228 pKa = 3.82WPQRR232 pKa = 11.84PSSSGLQHH240 pKa = 6.83EE241 pKa = 5.16SPFTPRR247 pKa = 11.84GHH249 pKa = 6.69GRR251 pKa = 11.84GCVRR255 pKa = 11.84GRR257 pKa = 11.84RR258 pKa = 11.84SSGSSPHH265 pKa = 5.4QRR267 pKa = 11.84GDD269 pKa = 2.76RR270 pKa = 11.84GGRR273 pKa = 11.84GRR275 pKa = 11.84SQSPSTPTTPGALPPDD291 pKa = 4.07LSFAGGDD298 pKa = 3.22RR299 pKa = 11.84SGGGGGGGGGGRR311 pKa = 11.84QRR313 pKa = 11.84GRR315 pKa = 11.84RR316 pKa = 11.84GSRR319 pKa = 11.84GPPRR323 pKa = 11.84LSPYY327 pKa = 9.95IPLGQVEE334 pKa = 4.43QGPGRR339 pKa = 11.84PEE341 pKa = 3.67GAALQRR347 pKa = 11.84PGRR350 pKa = 11.84PEE352 pKa = 3.59KK353 pKa = 10.75NPRR356 pKa = 11.84LSSTTPVVVLKK367 pKa = 11.19GPGNALKK374 pKa = 10.35CWRR377 pKa = 11.84LRR379 pKa = 11.84AKK381 pKa = 10.1AKK383 pKa = 10.45HH384 pKa = 5.65GALFCAISTAFSWVEE399 pKa = 3.66KK400 pKa = 10.25SSSSRR405 pKa = 11.84IGRR408 pKa = 11.84HH409 pKa = 5.7RR410 pKa = 11.84ILVGFINEE418 pKa = 4.0EE419 pKa = 3.71QRR421 pKa = 11.84EE422 pKa = 4.23DD423 pKa = 3.46FLRR426 pKa = 11.84TVRR429 pKa = 11.84LPRR432 pKa = 11.84GVEE435 pKa = 4.12CVPGGLDD442 pKa = 3.5SLL444 pKa = 4.5
Molecular weight: 49.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2507 |
101 |
608 |
358.1 |
39.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.465 ± 0.372 | 2.553 ± 0.826 |
5.704 ± 0.53 | 6.462 ± 0.469 |
4.108 ± 0.625 | 8.935 ± 1.523 |
1.635 ± 0.159 | 3.47 ± 0.348 |
4.826 ± 0.805 | 8.895 ± 0.712 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.715 ± 0.268 | 3.391 ± 0.592 |
7.06 ± 1.106 | 4.906 ± 0.676 |
6.223 ± 0.751 | 8.496 ± 0.657 |
5.744 ± 0.559 | 6.223 ± 0.586 |
1.316 ± 0.344 | 2.832 ± 0.514 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
