
Hubei virga-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
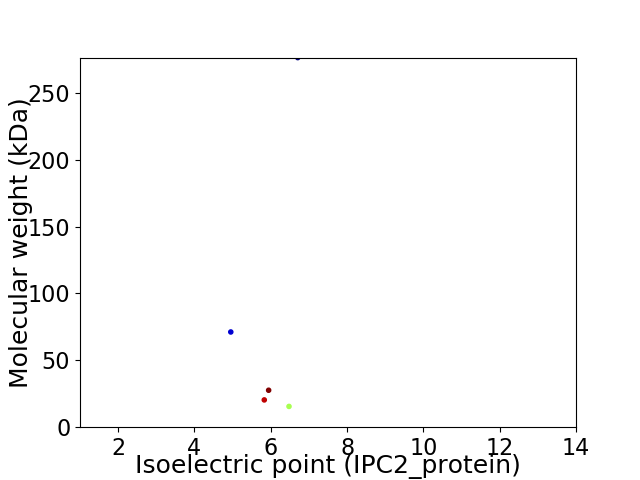
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJT5|A0A1L3KJT5_9VIRU Uncharacterized protein OS=Hubei virga-like virus 18 OX=1923333 PE=4 SV=1
MM1 pKa = 7.63RR2 pKa = 11.84LIIFVLFLFFLYY14 pKa = 10.21IPVHH18 pKa = 6.61SYY20 pKa = 6.78TTNPTSLRR28 pKa = 11.84QVCCQCRR35 pKa = 11.84DD36 pKa = 3.18RR37 pKa = 11.84SLTCNFCTQYY47 pKa = 10.68TDD49 pKa = 3.55SHH51 pKa = 6.95GCDD54 pKa = 3.11LSGSQLTTWAISVSTYY70 pKa = 10.67FEE72 pKa = 4.37TNLIPTHH79 pKa = 6.23TSTFASSYY87 pKa = 10.22GYY89 pKa = 10.95LYY91 pKa = 10.37FLKK94 pKa = 10.62DD95 pKa = 3.59SIVTSFAAAAPSCSRR110 pKa = 11.84HH111 pKa = 5.45TNTAQYY117 pKa = 10.81LYY119 pKa = 10.64GDD121 pKa = 5.24SILKK125 pKa = 10.58DD126 pKa = 3.89YY127 pKa = 10.63DD128 pKa = 3.71CSSATVGSSCGVGRR142 pKa = 11.84YY143 pKa = 8.74CSSAITTTSGDD154 pKa = 3.0THH156 pKa = 7.19YY157 pKa = 11.09CFSQYY162 pKa = 11.39AKK164 pKa = 10.32LPSIEE169 pKa = 4.26WYY171 pKa = 10.22DD172 pKa = 3.13INYY175 pKa = 9.89YY176 pKa = 8.78YY177 pKa = 11.22NKK179 pKa = 9.82LASPTGQCVEE189 pKa = 4.09ANYY192 pKa = 10.05PSLLSAMKK200 pKa = 9.67EE201 pKa = 4.01LSSAPASSVTVNGYY215 pKa = 10.75DD216 pKa = 3.61DD217 pKa = 3.95TSMQISVRR225 pKa = 11.84STDD228 pKa = 3.5YY229 pKa = 11.53LLNTYY234 pKa = 10.74RR235 pKa = 11.84MLEE238 pKa = 3.99RR239 pKa = 11.84GLPFAKK245 pKa = 9.04RR246 pKa = 11.84TYY248 pKa = 10.96QNIEE252 pKa = 4.27TYY254 pKa = 9.39CTDD257 pKa = 3.55DD258 pKa = 3.5PVVTSRR264 pKa = 11.84IFSSADD270 pKa = 3.22QMYY273 pKa = 8.2TWYY276 pKa = 11.09LNVLVIRR283 pKa = 11.84RR284 pKa = 11.84GPCSLYY290 pKa = 10.55VPYY293 pKa = 10.79SEE295 pKa = 5.65AQDD298 pKa = 3.19ICADD302 pKa = 3.9DD303 pKa = 5.43LLISDD308 pKa = 4.54YY309 pKa = 10.47TASCPDD315 pKa = 3.18SFRR318 pKa = 11.84PRR320 pKa = 11.84QFSPNRR326 pKa = 11.84WLDD329 pKa = 3.53YY330 pKa = 10.99NITVPNPNSTCDD342 pKa = 3.09KK343 pKa = 9.48MKK345 pKa = 10.67EE346 pKa = 3.92LFEE349 pKa = 5.4CGVKK353 pKa = 10.37DD354 pKa = 3.83WIDD357 pKa = 3.36YY358 pKa = 7.9FANLTAGGGSGGSANYY374 pKa = 9.54EE375 pKa = 3.61EE376 pKa = 5.03HH377 pKa = 7.36ARR379 pKa = 11.84LTDD382 pKa = 3.67EE383 pKa = 4.25YY384 pKa = 11.27AQARR388 pKa = 11.84QFGWFSWISSVFEE401 pKa = 4.4PGIKK405 pKa = 10.27LIFDD409 pKa = 3.82IFGSNFEE416 pKa = 4.85DD417 pKa = 4.17YY418 pKa = 11.19VVTFFEE424 pKa = 4.95KK425 pKa = 10.43LLEE428 pKa = 4.46YY429 pKa = 10.19ILQIVFEE436 pKa = 4.81LFNSIVALFKK446 pKa = 10.7KK447 pKa = 10.0SQQFIDD453 pKa = 3.65KK454 pKa = 9.91LVTFITRR461 pKa = 11.84ILDD464 pKa = 3.39VLFSFIAFLLKK475 pKa = 10.73ALIGVVLKK483 pKa = 10.7IEE485 pKa = 3.78QHH487 pKa = 5.59YY488 pKa = 11.14LLFEE492 pKa = 4.16YY493 pKa = 10.83VLLFLLVDD501 pKa = 3.88YY502 pKa = 10.81YY503 pKa = 11.34LINNNIFSLLVVLLVMVVVGIDD525 pKa = 3.18RR526 pKa = 11.84RR527 pKa = 11.84SPSILLAFHH536 pKa = 6.27SLEE539 pKa = 3.91YY540 pKa = 10.56AYY542 pKa = 11.36VNLSGYY548 pKa = 10.37DD549 pKa = 3.73PSSLTWDD556 pKa = 3.48YY557 pKa = 11.66SLTYY561 pKa = 10.22HH562 pKa = 6.61SYY564 pKa = 11.11SRR566 pKa = 11.84NKK568 pKa = 8.85TYY570 pKa = 10.77NISFPPLPEE579 pKa = 5.5LPDD582 pKa = 3.34VPLYY586 pKa = 9.01NTSTQVNHH594 pKa = 7.01TYY596 pKa = 10.51PLYY599 pKa = 10.17EE600 pKa = 4.43IKK602 pKa = 10.71NHH604 pKa = 5.75TVDD607 pKa = 3.63CDD609 pKa = 3.75SYY611 pKa = 11.23PLYY614 pKa = 9.84NTSSYY619 pKa = 10.65FIHH622 pKa = 6.4QAA624 pKa = 2.95
MM1 pKa = 7.63RR2 pKa = 11.84LIIFVLFLFFLYY14 pKa = 10.21IPVHH18 pKa = 6.61SYY20 pKa = 6.78TTNPTSLRR28 pKa = 11.84QVCCQCRR35 pKa = 11.84DD36 pKa = 3.18RR37 pKa = 11.84SLTCNFCTQYY47 pKa = 10.68TDD49 pKa = 3.55SHH51 pKa = 6.95GCDD54 pKa = 3.11LSGSQLTTWAISVSTYY70 pKa = 10.67FEE72 pKa = 4.37TNLIPTHH79 pKa = 6.23TSTFASSYY87 pKa = 10.22GYY89 pKa = 10.95LYY91 pKa = 10.37FLKK94 pKa = 10.62DD95 pKa = 3.59SIVTSFAAAAPSCSRR110 pKa = 11.84HH111 pKa = 5.45TNTAQYY117 pKa = 10.81LYY119 pKa = 10.64GDD121 pKa = 5.24SILKK125 pKa = 10.58DD126 pKa = 3.89YY127 pKa = 10.63DD128 pKa = 3.71CSSATVGSSCGVGRR142 pKa = 11.84YY143 pKa = 8.74CSSAITTTSGDD154 pKa = 3.0THH156 pKa = 7.19YY157 pKa = 11.09CFSQYY162 pKa = 11.39AKK164 pKa = 10.32LPSIEE169 pKa = 4.26WYY171 pKa = 10.22DD172 pKa = 3.13INYY175 pKa = 9.89YY176 pKa = 8.78YY177 pKa = 11.22NKK179 pKa = 9.82LASPTGQCVEE189 pKa = 4.09ANYY192 pKa = 10.05PSLLSAMKK200 pKa = 9.67EE201 pKa = 4.01LSSAPASSVTVNGYY215 pKa = 10.75DD216 pKa = 3.61DD217 pKa = 3.95TSMQISVRR225 pKa = 11.84STDD228 pKa = 3.5YY229 pKa = 11.53LLNTYY234 pKa = 10.74RR235 pKa = 11.84MLEE238 pKa = 3.99RR239 pKa = 11.84GLPFAKK245 pKa = 9.04RR246 pKa = 11.84TYY248 pKa = 10.96QNIEE252 pKa = 4.27TYY254 pKa = 9.39CTDD257 pKa = 3.55DD258 pKa = 3.5PVVTSRR264 pKa = 11.84IFSSADD270 pKa = 3.22QMYY273 pKa = 8.2TWYY276 pKa = 11.09LNVLVIRR283 pKa = 11.84RR284 pKa = 11.84GPCSLYY290 pKa = 10.55VPYY293 pKa = 10.79SEE295 pKa = 5.65AQDD298 pKa = 3.19ICADD302 pKa = 3.9DD303 pKa = 5.43LLISDD308 pKa = 4.54YY309 pKa = 10.47TASCPDD315 pKa = 3.18SFRR318 pKa = 11.84PRR320 pKa = 11.84QFSPNRR326 pKa = 11.84WLDD329 pKa = 3.53YY330 pKa = 10.99NITVPNPNSTCDD342 pKa = 3.09KK343 pKa = 9.48MKK345 pKa = 10.67EE346 pKa = 3.92LFEE349 pKa = 5.4CGVKK353 pKa = 10.37DD354 pKa = 3.83WIDD357 pKa = 3.36YY358 pKa = 7.9FANLTAGGGSGGSANYY374 pKa = 9.54EE375 pKa = 3.61EE376 pKa = 5.03HH377 pKa = 7.36ARR379 pKa = 11.84LTDD382 pKa = 3.67EE383 pKa = 4.25YY384 pKa = 11.27AQARR388 pKa = 11.84QFGWFSWISSVFEE401 pKa = 4.4PGIKK405 pKa = 10.27LIFDD409 pKa = 3.82IFGSNFEE416 pKa = 4.85DD417 pKa = 4.17YY418 pKa = 11.19VVTFFEE424 pKa = 4.95KK425 pKa = 10.43LLEE428 pKa = 4.46YY429 pKa = 10.19ILQIVFEE436 pKa = 4.81LFNSIVALFKK446 pKa = 10.7KK447 pKa = 10.0SQQFIDD453 pKa = 3.65KK454 pKa = 9.91LVTFITRR461 pKa = 11.84ILDD464 pKa = 3.39VLFSFIAFLLKK475 pKa = 10.73ALIGVVLKK483 pKa = 10.7IEE485 pKa = 3.78QHH487 pKa = 5.59YY488 pKa = 11.14LLFEE492 pKa = 4.16YY493 pKa = 10.83VLLFLLVDD501 pKa = 3.88YY502 pKa = 10.81YY503 pKa = 11.34LINNNIFSLLVVLLVMVVVGIDD525 pKa = 3.18RR526 pKa = 11.84RR527 pKa = 11.84SPSILLAFHH536 pKa = 6.27SLEE539 pKa = 3.91YY540 pKa = 10.56AYY542 pKa = 11.36VNLSGYY548 pKa = 10.37DD549 pKa = 3.73PSSLTWDD556 pKa = 3.48YY557 pKa = 11.66SLTYY561 pKa = 10.22HH562 pKa = 6.61SYY564 pKa = 11.11SRR566 pKa = 11.84NKK568 pKa = 8.85TYY570 pKa = 10.77NISFPPLPEE579 pKa = 5.5LPDD582 pKa = 3.34VPLYY586 pKa = 9.01NTSTQVNHH594 pKa = 7.01TYY596 pKa = 10.51PLYY599 pKa = 10.17EE600 pKa = 4.43IKK602 pKa = 10.71NHH604 pKa = 5.75TVDD607 pKa = 3.63CDD609 pKa = 3.75SYY611 pKa = 11.23PLYY614 pKa = 9.84NTSSYY619 pKa = 10.65FIHH622 pKa = 6.4QAA624 pKa = 2.95
Molecular weight: 71.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJZ2|A0A1L3KJZ2_9VIRU Uncharacterized protein OS=Hubei virga-like virus 18 OX=1923333 PE=4 SV=1
MM1 pKa = 7.67PNIPASILYY10 pKa = 10.27DD11 pKa = 3.41FVVCSLKK18 pKa = 9.39TQVDD22 pKa = 3.1IAKK25 pKa = 9.9RR26 pKa = 11.84VRR28 pKa = 11.84DD29 pKa = 3.96IQNADD34 pKa = 3.86YY35 pKa = 11.1ILFDD39 pKa = 3.78PSFRR43 pKa = 11.84RR44 pKa = 11.84THH46 pKa = 7.43RR47 pKa = 11.84IRR49 pKa = 11.84IGDD52 pKa = 3.48YY53 pKa = 9.89RR54 pKa = 11.84VEE56 pKa = 3.8VDD58 pKa = 3.03AMGRR62 pKa = 11.84KK63 pKa = 8.98AVTCAAAKK71 pKa = 10.03EE72 pKa = 4.3VFDD75 pKa = 5.14TDD77 pKa = 5.17DD78 pKa = 2.95YY79 pKa = 9.14WQIVRR84 pKa = 11.84EE85 pKa = 4.25FDD87 pKa = 3.44CDD89 pKa = 4.82DD90 pKa = 4.69IILHH94 pKa = 5.67HH95 pKa = 6.95HH96 pKa = 6.21CVALRR101 pKa = 11.84AKK103 pKa = 9.71PVVKK107 pKa = 10.38SRR109 pKa = 11.84SKK111 pKa = 10.13LTLDD115 pKa = 3.67IPPPTHH121 pKa = 6.28HH122 pKa = 7.04HH123 pKa = 6.41PASLSDD129 pKa = 3.16ATTPPSSPP137 pKa = 3.3
MM1 pKa = 7.67PNIPASILYY10 pKa = 10.27DD11 pKa = 3.41FVVCSLKK18 pKa = 9.39TQVDD22 pKa = 3.1IAKK25 pKa = 9.9RR26 pKa = 11.84VRR28 pKa = 11.84DD29 pKa = 3.96IQNADD34 pKa = 3.86YY35 pKa = 11.1ILFDD39 pKa = 3.78PSFRR43 pKa = 11.84RR44 pKa = 11.84THH46 pKa = 7.43RR47 pKa = 11.84IRR49 pKa = 11.84IGDD52 pKa = 3.48YY53 pKa = 9.89RR54 pKa = 11.84VEE56 pKa = 3.8VDD58 pKa = 3.03AMGRR62 pKa = 11.84KK63 pKa = 8.98AVTCAAAKK71 pKa = 10.03EE72 pKa = 4.3VFDD75 pKa = 5.14TDD77 pKa = 5.17DD78 pKa = 2.95YY79 pKa = 9.14WQIVRR84 pKa = 11.84EE85 pKa = 4.25FDD87 pKa = 3.44CDD89 pKa = 4.82DD90 pKa = 4.69IILHH94 pKa = 5.67HH95 pKa = 6.95HH96 pKa = 6.21CVALRR101 pKa = 11.84AKK103 pKa = 9.71PVVKK107 pKa = 10.38SRR109 pKa = 11.84SKK111 pKa = 10.13LTLDD115 pKa = 3.67IPPPTHH121 pKa = 6.28HH122 pKa = 7.04HH123 pKa = 6.41PASLSDD129 pKa = 3.16ATTPPSSPP137 pKa = 3.3
Molecular weight: 15.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3620 |
137 |
2424 |
724.0 |
82.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.022 ± 0.576 | 2.127 ± 0.27 |
6.713 ± 0.742 | 3.536 ± 0.342 |
5.497 ± 1.029 | 4.116 ± 0.381 |
2.762 ± 0.449 | 7.21 ± 0.374 |
4.53 ± 0.747 | 9.586 ± 0.634 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.602 ± 0.147 | 5.359 ± 0.556 |
4.834 ± 0.41 | 2.901 ± 0.222 |
5.138 ± 0.844 | 8.729 ± 1.019 |
6.547 ± 0.492 | 6.602 ± 0.501 |
1.022 ± 0.084 | 5.166 ± 0.877 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
