
Actinomadura sp. WAC 06369
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Thermomonosporaceae; Actinomadura; unclassified Actinomadura
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
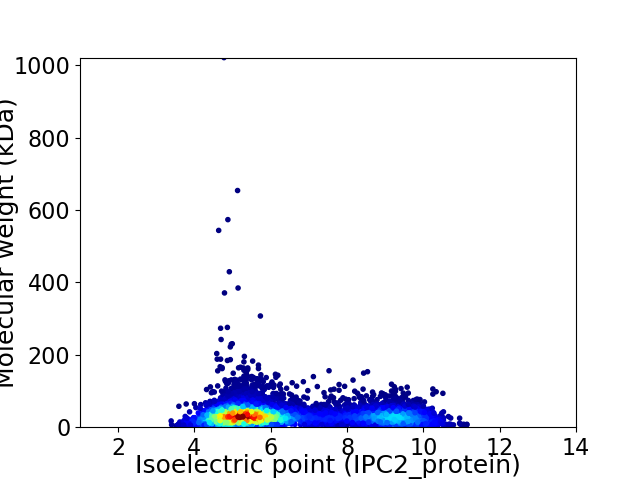
Virtual 2D-PAGE plot for 7670 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A429FJA4|A0A429FJA4_9ACTN Uncharacterized protein OS=Actinomadura sp. WAC 06369 OX=2203193 GN=DMH08_20880 PE=4 SV=1
MM1 pKa = 7.32VFGLLVVVVLVVVLVVLGLRR21 pKa = 11.84ASKK24 pKa = 10.56AGRR27 pKa = 11.84DD28 pKa = 3.64DD29 pKa = 5.11DD30 pKa = 5.54DD31 pKa = 4.14WMDD34 pKa = 4.8DD35 pKa = 3.64EE36 pKa = 4.89QPQPRR41 pKa = 11.84GRR43 pKa = 11.84RR44 pKa = 11.84GAAQQDD50 pKa = 4.26EE51 pKa = 4.87MNGPDD56 pKa = 4.51DD57 pKa = 3.92YY58 pKa = 11.65GYY60 pKa = 11.22DD61 pKa = 3.61DD62 pKa = 4.77GGYY65 pKa = 9.71GAPAPGPDD73 pKa = 3.32RR74 pKa = 11.84RR75 pKa = 11.84VAGAGAGAGGPLAAPAGQQGPPAPPPSGQASDD107 pKa = 3.99EE108 pKa = 4.36MEE110 pKa = 5.66DD111 pKa = 3.38DD112 pKa = 5.39DD113 pKa = 4.41YY114 pKa = 11.55WATITFDD121 pKa = 3.31KK122 pKa = 11.02PKK124 pKa = 10.51FPWQHH129 pKa = 4.71NQEE132 pKa = 4.2GQRR135 pKa = 11.84PDD137 pKa = 3.45GDD139 pKa = 3.83HH140 pKa = 7.05AADD143 pKa = 4.29PLHH146 pKa = 6.66AQGGDD151 pKa = 3.3DD152 pKa = 5.36GYY154 pKa = 11.53GDD156 pKa = 3.6QYY158 pKa = 11.89GDD160 pKa = 3.34QQYY163 pKa = 11.61GDD165 pKa = 3.67QQYY168 pKa = 11.58GDD170 pKa = 3.6QQYY173 pKa = 10.81GDD175 pKa = 3.98QYY177 pKa = 11.38ADD179 pKa = 3.14QYY181 pKa = 11.91GDD183 pKa = 3.23QQYY186 pKa = 11.62ADD188 pKa = 3.5QQQYY192 pKa = 9.92GDD194 pKa = 3.7QYY196 pKa = 10.1GDD198 pKa = 3.18QYY200 pKa = 11.85AEE202 pKa = 4.38QDD204 pKa = 3.83PQVPDD209 pKa = 3.1HH210 pKa = 6.76GMNQTVTLGQEE221 pKa = 4.09GGAFGAPGGPGQGPPNQGPPGPGGPGQTGGQPAPTGPQQAPGGPGGFQGGSPFGGGDD278 pKa = 3.49PGATALDD285 pKa = 5.26PIPADD290 pKa = 3.51LAGGPGMQGGPGAHH304 pKa = 6.74APFAPGGPDD313 pKa = 2.85QHH315 pKa = 8.63VYY317 pKa = 10.65GGQDD321 pKa = 3.13AQPAYY326 pKa = 10.42GSAEE330 pKa = 4.03PPFGGPEE337 pKa = 3.73QQPPYY342 pKa = 10.64GGDD345 pKa = 3.14RR346 pKa = 11.84QQPYY350 pKa = 10.7AEE352 pKa = 4.3PQQPGMPPEE361 pKa = 4.18PAPYY365 pKa = 10.17GLPEE369 pKa = 3.85QPQPYY374 pKa = 10.11GGDD377 pKa = 3.52PLASGGRR384 pKa = 11.84PTPASPPPGRR394 pKa = 11.84PDD396 pKa = 3.47AFDD399 pKa = 3.72MPLGASGPGSTGPNQGPGSTGPNQGPPAPPPSDD432 pKa = 3.69PLGLPLGRR440 pKa = 11.84TDD442 pKa = 4.13EE443 pKa = 4.37PAAPGGSGPDD453 pKa = 3.5TDD455 pKa = 3.54NHH457 pKa = 6.43KK458 pKa = 10.98LPTVDD463 pKa = 5.16EE464 pKa = 4.24LLQRR468 pKa = 11.84IQSDD472 pKa = 3.97RR473 pKa = 11.84QRR475 pKa = 11.84SSGPPSGDD483 pKa = 3.01SGGSSYY489 pKa = 11.15GGPLGDD495 pKa = 4.64PLSDD499 pKa = 3.72PLATGSYY506 pKa = 10.04GGSTGAGPGTGQGSSTGPWSPGGGLGNSGGYY537 pKa = 7.93PQGGQSEE544 pKa = 5.69GYY546 pKa = 8.35PASPSYY552 pKa = 11.58GEE554 pKa = 4.15GNRR557 pKa = 11.84YY558 pKa = 9.8DD559 pKa = 4.64DD560 pKa = 4.65PLSGGRR566 pKa = 11.84DD567 pKa = 3.37AYY569 pKa = 10.62GGEE572 pKa = 4.34SGSGGAGAYY581 pKa = 10.38GDD583 pKa = 4.49FSGSSYY589 pKa = 11.05SGNDD593 pKa = 3.77PLSAPHH599 pKa = 7.4DD600 pKa = 4.5PGAQSGDD607 pKa = 3.75PNSTQAYY614 pKa = 8.96DD615 pKa = 3.61PGFGRR620 pKa = 11.84QGGYY624 pKa = 10.04QGDD627 pKa = 3.59DD628 pKa = 3.34WEE630 pKa = 4.16NHH632 pKa = 4.55RR633 pKa = 11.84DD634 pKa = 3.59YY635 pKa = 11.29RR636 pKa = 11.84RR637 pKa = 3.16
MM1 pKa = 7.32VFGLLVVVVLVVVLVVLGLRR21 pKa = 11.84ASKK24 pKa = 10.56AGRR27 pKa = 11.84DD28 pKa = 3.64DD29 pKa = 5.11DD30 pKa = 5.54DD31 pKa = 4.14WMDD34 pKa = 4.8DD35 pKa = 3.64EE36 pKa = 4.89QPQPRR41 pKa = 11.84GRR43 pKa = 11.84RR44 pKa = 11.84GAAQQDD50 pKa = 4.26EE51 pKa = 4.87MNGPDD56 pKa = 4.51DD57 pKa = 3.92YY58 pKa = 11.65GYY60 pKa = 11.22DD61 pKa = 3.61DD62 pKa = 4.77GGYY65 pKa = 9.71GAPAPGPDD73 pKa = 3.32RR74 pKa = 11.84RR75 pKa = 11.84VAGAGAGAGGPLAAPAGQQGPPAPPPSGQASDD107 pKa = 3.99EE108 pKa = 4.36MEE110 pKa = 5.66DD111 pKa = 3.38DD112 pKa = 5.39DD113 pKa = 4.41YY114 pKa = 11.55WATITFDD121 pKa = 3.31KK122 pKa = 11.02PKK124 pKa = 10.51FPWQHH129 pKa = 4.71NQEE132 pKa = 4.2GQRR135 pKa = 11.84PDD137 pKa = 3.45GDD139 pKa = 3.83HH140 pKa = 7.05AADD143 pKa = 4.29PLHH146 pKa = 6.66AQGGDD151 pKa = 3.3DD152 pKa = 5.36GYY154 pKa = 11.53GDD156 pKa = 3.6QYY158 pKa = 11.89GDD160 pKa = 3.34QQYY163 pKa = 11.61GDD165 pKa = 3.67QQYY168 pKa = 11.58GDD170 pKa = 3.6QQYY173 pKa = 10.81GDD175 pKa = 3.98QYY177 pKa = 11.38ADD179 pKa = 3.14QYY181 pKa = 11.91GDD183 pKa = 3.23QQYY186 pKa = 11.62ADD188 pKa = 3.5QQQYY192 pKa = 9.92GDD194 pKa = 3.7QYY196 pKa = 10.1GDD198 pKa = 3.18QYY200 pKa = 11.85AEE202 pKa = 4.38QDD204 pKa = 3.83PQVPDD209 pKa = 3.1HH210 pKa = 6.76GMNQTVTLGQEE221 pKa = 4.09GGAFGAPGGPGQGPPNQGPPGPGGPGQTGGQPAPTGPQQAPGGPGGFQGGSPFGGGDD278 pKa = 3.49PGATALDD285 pKa = 5.26PIPADD290 pKa = 3.51LAGGPGMQGGPGAHH304 pKa = 6.74APFAPGGPDD313 pKa = 2.85QHH315 pKa = 8.63VYY317 pKa = 10.65GGQDD321 pKa = 3.13AQPAYY326 pKa = 10.42GSAEE330 pKa = 4.03PPFGGPEE337 pKa = 3.73QQPPYY342 pKa = 10.64GGDD345 pKa = 3.14RR346 pKa = 11.84QQPYY350 pKa = 10.7AEE352 pKa = 4.3PQQPGMPPEE361 pKa = 4.18PAPYY365 pKa = 10.17GLPEE369 pKa = 3.85QPQPYY374 pKa = 10.11GGDD377 pKa = 3.52PLASGGRR384 pKa = 11.84PTPASPPPGRR394 pKa = 11.84PDD396 pKa = 3.47AFDD399 pKa = 3.72MPLGASGPGSTGPNQGPGSTGPNQGPPAPPPSDD432 pKa = 3.69PLGLPLGRR440 pKa = 11.84TDD442 pKa = 4.13EE443 pKa = 4.37PAAPGGSGPDD453 pKa = 3.5TDD455 pKa = 3.54NHH457 pKa = 6.43KK458 pKa = 10.98LPTVDD463 pKa = 5.16EE464 pKa = 4.24LLQRR468 pKa = 11.84IQSDD472 pKa = 3.97RR473 pKa = 11.84QRR475 pKa = 11.84SSGPPSGDD483 pKa = 3.01SGGSSYY489 pKa = 11.15GGPLGDD495 pKa = 4.64PLSDD499 pKa = 3.72PLATGSYY506 pKa = 10.04GGSTGAGPGTGQGSSTGPWSPGGGLGNSGGYY537 pKa = 7.93PQGGQSEE544 pKa = 5.69GYY546 pKa = 8.35PASPSYY552 pKa = 11.58GEE554 pKa = 4.15GNRR557 pKa = 11.84YY558 pKa = 9.8DD559 pKa = 4.64DD560 pKa = 4.65PLSGGRR566 pKa = 11.84DD567 pKa = 3.37AYY569 pKa = 10.62GGEE572 pKa = 4.34SGSGGAGAYY581 pKa = 10.38GDD583 pKa = 4.49FSGSSYY589 pKa = 11.05SGNDD593 pKa = 3.77PLSAPHH599 pKa = 7.4DD600 pKa = 4.5PGAQSGDD607 pKa = 3.75PNSTQAYY614 pKa = 8.96DD615 pKa = 3.61PGFGRR620 pKa = 11.84QGGYY624 pKa = 10.04QGDD627 pKa = 3.59DD628 pKa = 3.34WEE630 pKa = 4.16NHH632 pKa = 4.55RR633 pKa = 11.84DD634 pKa = 3.59YY635 pKa = 11.29RR636 pKa = 11.84RR637 pKa = 3.16
Molecular weight: 64.09 kDa
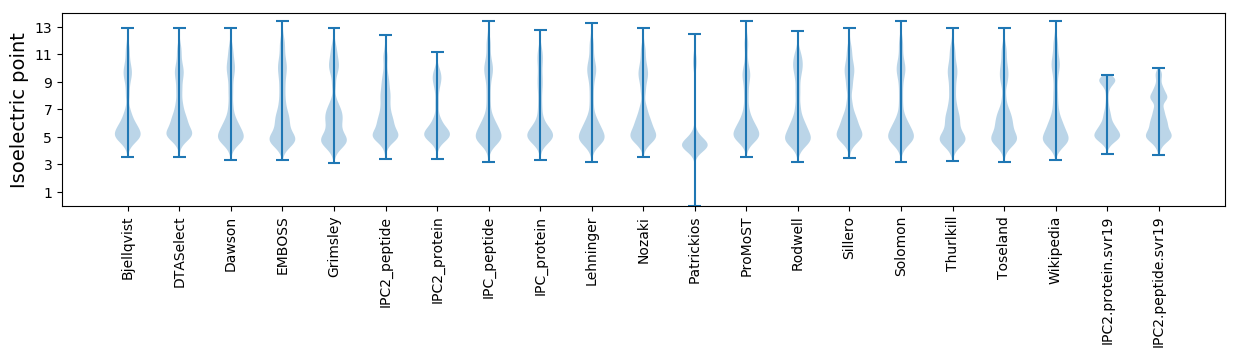
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A429FF43|A0A429FF43_9ACTN CoA transferase OS=Actinomadura sp. WAC 06369 OX=2203193 GN=DMH08_21255 PE=4 SV=1
SS1 pKa = 7.08RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84CGRR7 pKa = 11.84GGTARR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84RR17 pKa = 11.84GRR19 pKa = 11.84GARR22 pKa = 11.84RR23 pKa = 11.84GRR25 pKa = 11.84RR26 pKa = 11.84VRR28 pKa = 11.84AGRR31 pKa = 11.84GRR33 pKa = 11.84RR34 pKa = 11.84GRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84GGAGPGGRR47 pKa = 11.84AGRR50 pKa = 11.84AARR53 pKa = 11.84GGGRR57 pKa = 11.84DD58 pKa = 3.16RR59 pKa = 11.84RR60 pKa = 3.94
SS1 pKa = 7.08RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84CGRR7 pKa = 11.84GGTARR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84RR17 pKa = 11.84GRR19 pKa = 11.84GARR22 pKa = 11.84RR23 pKa = 11.84GRR25 pKa = 11.84RR26 pKa = 11.84VRR28 pKa = 11.84AGRR31 pKa = 11.84GRR33 pKa = 11.84RR34 pKa = 11.84GRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84GGAGPGGRR47 pKa = 11.84AGRR50 pKa = 11.84AARR53 pKa = 11.84GGGRR57 pKa = 11.84DD58 pKa = 3.16RR59 pKa = 11.84RR60 pKa = 3.94
Molecular weight: 6.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2493908 |
18 |
9614 |
325.2 |
34.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.655 ± 0.044 | 0.749 ± 0.008 |
6.363 ± 0.025 | 5.771 ± 0.023 |
2.769 ± 0.017 | 9.944 ± 0.026 |
2.141 ± 0.013 | 3.039 ± 0.019 |
1.73 ± 0.019 | 10.207 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.798 ± 0.011 | 1.541 ± 0.013 |
6.394 ± 0.026 | 2.137 ± 0.014 |
9.048 ± 0.031 | 4.278 ± 0.018 |
5.326 ± 0.023 | 8.716 ± 0.03 |
1.474 ± 0.013 | 1.922 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
