
Verticillium dahliae (strain VdLs.17 / ATCC MYA-4575 / FGSC 10137) (Verticillium wilt)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Glomerellales; Plectosphaerellaceae; Verticillium; Verticillium dahliae
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
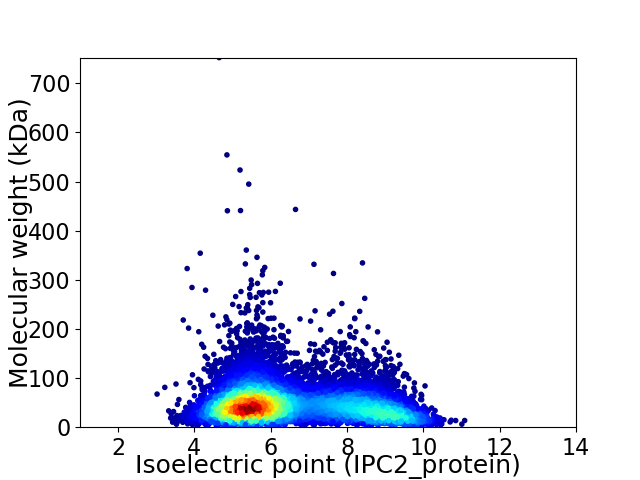
Virtual 2D-PAGE plot for 10530 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2X2Y2|G2X2Y2_VERDV Uncharacterized protein OS=Verticillium dahliae (strain VdLs.17 / ATCC MYA-4575 / FGSC 10137) OX=498257 GN=VDAG_04176 PE=4 SV=1
MM1 pKa = 7.12PQALRR6 pKa = 11.84VLPVALLAARR16 pKa = 11.84LPDD19 pKa = 3.8LSFLKK24 pKa = 10.59SLPGLVNEE32 pKa = 5.55AGPGDD37 pKa = 4.2PGAGAGTGPGTIIPDD52 pKa = 3.75AVNSIPGVIGGALGNGPASGIVPGLANALPGLVNDD87 pKa = 4.98TLAGGPGSGPGTLLPDD103 pKa = 3.65NVNNIPSVIGGALGNSTTSAGLIPGLIGSLPGIVNDD139 pKa = 4.97TIGGAAPGPLIPDD152 pKa = 4.24AVNNMPSIIGGAIGSVPGNGSAIVPGLLNDD182 pKa = 4.72LPGLVNGVFQNQTGAGTVLPDD203 pKa = 5.83AINTLPGVIGGALSGDD219 pKa = 3.88NPGSAIPTVIDD230 pKa = 3.95NLPGLINGSVGQTAPDD246 pKa = 3.18SGGAILPGPLNEE258 pKa = 4.55LPGIIGGAVNGSTPSPEE275 pKa = 3.84GVVPGLVNNLPAIVAGASGNQSGAGSILPGPLNEE309 pKa = 5.72LPGLLNSTLGAGGPVAGTLVPGIVNGLPDD338 pKa = 4.64LVNGALNGTNGGSSAGPGGLPGTLLPGGLNEE369 pKa = 5.2LPGNLTAGLPNGTTAGTLLPSIIGGLPGAVNSVVNGTAGTLIPGVVNQVPGIIKK423 pKa = 10.2GAVDD427 pKa = 3.85SGPNIGPLIPGIVGSFPAIVNGTVSGGDD455 pKa = 3.29QPQQDD460 pKa = 5.22LEE462 pKa = 4.31TLIPGIIEE470 pKa = 4.43TIPTCVEE477 pKa = 3.82GALGNVPNVNPADD490 pKa = 4.3IIPIVIGRR498 pKa = 11.84LPDD501 pKa = 4.5IISQLPNVTTPGQVLPGPLNQLPGIISGAISDD533 pKa = 4.12AAGVQPGVIAPGLVATLPSVLEE555 pKa = 3.98GLVNNLPNTGGAVRR569 pKa = 11.84PTIPGLSEE577 pKa = 4.22SLPGLCEE584 pKa = 3.99DD585 pKa = 4.76ALNSGSGALIPEE597 pKa = 4.55VVNTLPGVIGGAISNGQLPGSAISLPDD624 pKa = 3.78LVNNLPGLINEE635 pKa = 4.88ALNKK639 pKa = 9.77GQEE642 pKa = 3.98EE643 pKa = 5.19CIPGPVNNLPTTIGNVISNGSLPIPNLPISDD674 pKa = 4.65IINNLPEE681 pKa = 5.39LINGAIQTGAGTLIPDD697 pKa = 4.44RR698 pKa = 11.84LNNLPADD705 pKa = 3.53IGVSLNATVGLPVPEE720 pKa = 4.3ILANLPSIINNAVSSGNPVAIPEE743 pKa = 4.45VINNLPATIGTAVGGATNLIPQIIGNLSALVNEE776 pKa = 5.86AINSGASTVLPPVINNLPVTIGGALNGTTLPPAIISNLPALINGAIGNGSEE827 pKa = 4.36TAGGAATLLPEE838 pKa = 5.94AINTLPAVIGGSLGGLTGNAASLVPAIVANLPGLVTGALQSGAGTLVPPSVNNLPTTIGEE898 pKa = 4.4SLGNLTGNAGALVPEE913 pKa = 5.57IINNLPGLIAGGLVGGTGTLLPPAVNNLPAVIGGALGNVTGAAEE957 pKa = 4.11SLVPQIIANLPGLITGAIGSNSTTLIPDD985 pKa = 3.73VVNTLPVVIGGALTNSSIGFNPSIINNLPSLVNNAISGGNTTIIPPQVNNIPAVISGAIANGTTAGDD1052 pKa = 4.87LINNLIGNGTATAGGLLPILSGTLNGNVSIGGSSQGVLPGLIANVSVDD1100 pKa = 3.44ALGPNIPTLINGLIGTNGTLNAIAGGLLGNGTAGLTIPDD1139 pKa = 3.7ILGGIAAGTGNSSLPGLVGEE1159 pKa = 4.61ILGNGTGGGIIGGGGIGGAGAGLGVGVTVGGGLGAGGLLQNITTGLIGGGSGGASIPGVLPGIVAAGPRR1228 pKa = 11.84ANTTVPSVEE1237 pKa = 4.05VVLDD1241 pKa = 3.72NLGLSNLTASLPALVDD1257 pKa = 4.19SIIGSANLTGKK1268 pKa = 10.38NITGPAASLIDD1279 pKa = 3.81GLLGPGASAGSNVTLGPASGPGAGLIPNLLDD1310 pKa = 3.65NVATGNVTQIIPDD1323 pKa = 4.25LVNGLLGNGTANSGGLIPDD1342 pKa = 4.65LLQNASAGALPAVIPNIVGGLLGGATVGGAAGDD1375 pKa = 4.01LNNPGVGVNGSTNVNGTAGIGGILPTLVGSLNSGNLSNIVPGILTGLLGNGTASSGGLIPAVVVNINNGTTGANANANANATADD1459 pKa = 3.72VVNTGIIDD1467 pKa = 3.91GLLGAGSGGALLNTGGLLSGTRR1489 pKa = 11.84NLLTDD1494 pKa = 4.01SLNCGAIGIVCQGIQICRR1512 pKa = 11.84AGVCINSSGPSGTSTIPAAGTPTSPAAGISTTVVPGQGVVDD1553 pKa = 4.9LLNDD1557 pKa = 3.6SLNCGRR1563 pKa = 11.84LLNVCLFGQICSNGQCIVAGATTTAPPAAAQPTTTALPATTTVAGGNIINFLTDD1617 pKa = 3.23NLNCGRR1623 pKa = 11.84LLNVCLFGQVCRR1635 pKa = 11.84EE1636 pKa = 4.16GQCVSAAQATTAAIAPATTTIAVQVTTTAAGGQLTTVQVPTTVVAAAPTAAGADD1690 pKa = 3.45AAVGIVDD1697 pKa = 4.88HH1698 pKa = 7.07LNDD1701 pKa = 3.71SLNCGRR1707 pKa = 11.84LFNIYY1712 pKa = 10.05ILGQTCRR1719 pKa = 11.84NGVCVASGAAVPTTTAGTTAPTTTARR1745 pKa = 11.84ATTTAPVVGTTTTAAAAAPTTTVAAAPTTTAGGAFGVVDD1784 pKa = 5.21LLNDD1788 pKa = 3.76ALNCGRR1794 pKa = 11.84LLNICLFGQVCRR1806 pKa = 11.84SGVCVAAGAAVPTTTAPATTAPTTTLAATTAVVTTTLLTATRR1848 pKa = 11.84AATTTTVAGVPTTIAGTTTIAATITAAVAPTTTTGAATATTTGGTLAVSLDD1899 pKa = 3.59LHH1901 pKa = 5.92TDD1903 pKa = 3.29RR1904 pKa = 11.84LNCGRR1909 pKa = 11.84IGNICLGSQTCSSGLCISVGGAVVGGNTIATTATAATTTTTSTTAPAAAAPTTTAGTTTRR1969 pKa = 11.84AATTTTAAAPTTTAAAPTTTAAAPTTTGAQTNVAIAVSTTPTTAPAAVPTTTARR2023 pKa = 11.84VTTTVPTTPTVQAVPTTPTVQAVPTTPTVQAAPTTVRR2060 pKa = 11.84AATTTAAAPTTAPQAATTTQRR2081 pKa = 11.84AATTTAPSTGSSGSTGSSGSGASCGGFFNLIPCIFPLTCVNGRR2124 pKa = 11.84CQLLGVIGG2132 pKa = 3.74
MM1 pKa = 7.12PQALRR6 pKa = 11.84VLPVALLAARR16 pKa = 11.84LPDD19 pKa = 3.8LSFLKK24 pKa = 10.59SLPGLVNEE32 pKa = 5.55AGPGDD37 pKa = 4.2PGAGAGTGPGTIIPDD52 pKa = 3.75AVNSIPGVIGGALGNGPASGIVPGLANALPGLVNDD87 pKa = 4.98TLAGGPGSGPGTLLPDD103 pKa = 3.65NVNNIPSVIGGALGNSTTSAGLIPGLIGSLPGIVNDD139 pKa = 4.97TIGGAAPGPLIPDD152 pKa = 4.24AVNNMPSIIGGAIGSVPGNGSAIVPGLLNDD182 pKa = 4.72LPGLVNGVFQNQTGAGTVLPDD203 pKa = 5.83AINTLPGVIGGALSGDD219 pKa = 3.88NPGSAIPTVIDD230 pKa = 3.95NLPGLINGSVGQTAPDD246 pKa = 3.18SGGAILPGPLNEE258 pKa = 4.55LPGIIGGAVNGSTPSPEE275 pKa = 3.84GVVPGLVNNLPAIVAGASGNQSGAGSILPGPLNEE309 pKa = 5.72LPGLLNSTLGAGGPVAGTLVPGIVNGLPDD338 pKa = 4.64LVNGALNGTNGGSSAGPGGLPGTLLPGGLNEE369 pKa = 5.2LPGNLTAGLPNGTTAGTLLPSIIGGLPGAVNSVVNGTAGTLIPGVVNQVPGIIKK423 pKa = 10.2GAVDD427 pKa = 3.85SGPNIGPLIPGIVGSFPAIVNGTVSGGDD455 pKa = 3.29QPQQDD460 pKa = 5.22LEE462 pKa = 4.31TLIPGIIEE470 pKa = 4.43TIPTCVEE477 pKa = 3.82GALGNVPNVNPADD490 pKa = 4.3IIPIVIGRR498 pKa = 11.84LPDD501 pKa = 4.5IISQLPNVTTPGQVLPGPLNQLPGIISGAISDD533 pKa = 4.12AAGVQPGVIAPGLVATLPSVLEE555 pKa = 3.98GLVNNLPNTGGAVRR569 pKa = 11.84PTIPGLSEE577 pKa = 4.22SLPGLCEE584 pKa = 3.99DD585 pKa = 4.76ALNSGSGALIPEE597 pKa = 4.55VVNTLPGVIGGAISNGQLPGSAISLPDD624 pKa = 3.78LVNNLPGLINEE635 pKa = 4.88ALNKK639 pKa = 9.77GQEE642 pKa = 3.98EE643 pKa = 5.19CIPGPVNNLPTTIGNVISNGSLPIPNLPISDD674 pKa = 4.65IINNLPEE681 pKa = 5.39LINGAIQTGAGTLIPDD697 pKa = 4.44RR698 pKa = 11.84LNNLPADD705 pKa = 3.53IGVSLNATVGLPVPEE720 pKa = 4.3ILANLPSIINNAVSSGNPVAIPEE743 pKa = 4.45VINNLPATIGTAVGGATNLIPQIIGNLSALVNEE776 pKa = 5.86AINSGASTVLPPVINNLPVTIGGALNGTTLPPAIISNLPALINGAIGNGSEE827 pKa = 4.36TAGGAATLLPEE838 pKa = 5.94AINTLPAVIGGSLGGLTGNAASLVPAIVANLPGLVTGALQSGAGTLVPPSVNNLPTTIGEE898 pKa = 4.4SLGNLTGNAGALVPEE913 pKa = 5.57IINNLPGLIAGGLVGGTGTLLPPAVNNLPAVIGGALGNVTGAAEE957 pKa = 4.11SLVPQIIANLPGLITGAIGSNSTTLIPDD985 pKa = 3.73VVNTLPVVIGGALTNSSIGFNPSIINNLPSLVNNAISGGNTTIIPPQVNNIPAVISGAIANGTTAGDD1052 pKa = 4.87LINNLIGNGTATAGGLLPILSGTLNGNVSIGGSSQGVLPGLIANVSVDD1100 pKa = 3.44ALGPNIPTLINGLIGTNGTLNAIAGGLLGNGTAGLTIPDD1139 pKa = 3.7ILGGIAAGTGNSSLPGLVGEE1159 pKa = 4.61ILGNGTGGGIIGGGGIGGAGAGLGVGVTVGGGLGAGGLLQNITTGLIGGGSGGASIPGVLPGIVAAGPRR1228 pKa = 11.84ANTTVPSVEE1237 pKa = 4.05VVLDD1241 pKa = 3.72NLGLSNLTASLPALVDD1257 pKa = 4.19SIIGSANLTGKK1268 pKa = 10.38NITGPAASLIDD1279 pKa = 3.81GLLGPGASAGSNVTLGPASGPGAGLIPNLLDD1310 pKa = 3.65NVATGNVTQIIPDD1323 pKa = 4.25LVNGLLGNGTANSGGLIPDD1342 pKa = 4.65LLQNASAGALPAVIPNIVGGLLGGATVGGAAGDD1375 pKa = 4.01LNNPGVGVNGSTNVNGTAGIGGILPTLVGSLNSGNLSNIVPGILTGLLGNGTASSGGLIPAVVVNINNGTTGANANANANATADD1459 pKa = 3.72VVNTGIIDD1467 pKa = 3.91GLLGAGSGGALLNTGGLLSGTRR1489 pKa = 11.84NLLTDD1494 pKa = 4.01SLNCGAIGIVCQGIQICRR1512 pKa = 11.84AGVCINSSGPSGTSTIPAAGTPTSPAAGISTTVVPGQGVVDD1553 pKa = 4.9LLNDD1557 pKa = 3.6SLNCGRR1563 pKa = 11.84LLNVCLFGQICSNGQCIVAGATTTAPPAAAQPTTTALPATTTVAGGNIINFLTDD1617 pKa = 3.23NLNCGRR1623 pKa = 11.84LLNVCLFGQVCRR1635 pKa = 11.84EE1636 pKa = 4.16GQCVSAAQATTAAIAPATTTIAVQVTTTAAGGQLTTVQVPTTVVAAAPTAAGADD1690 pKa = 3.45AAVGIVDD1697 pKa = 4.88HH1698 pKa = 7.07LNDD1701 pKa = 3.71SLNCGRR1707 pKa = 11.84LFNIYY1712 pKa = 10.05ILGQTCRR1719 pKa = 11.84NGVCVASGAAVPTTTAGTTAPTTTARR1745 pKa = 11.84ATTTAPVVGTTTTAAAAAPTTTVAAAPTTTAGGAFGVVDD1784 pKa = 5.21LLNDD1788 pKa = 3.76ALNCGRR1794 pKa = 11.84LLNICLFGQVCRR1806 pKa = 11.84SGVCVAAGAAVPTTTAPATTAPTTTLAATTAVVTTTLLTATRR1848 pKa = 11.84AATTTTVAGVPTTIAGTTTIAATITAAVAPTTTTGAATATTTGGTLAVSLDD1899 pKa = 3.59LHH1901 pKa = 5.92TDD1903 pKa = 3.29RR1904 pKa = 11.84LNCGRR1909 pKa = 11.84IGNICLGSQTCSSGLCISVGGAVVGGNTIATTATAATTTTTSTTAPAAAAPTTTAGTTTRR1969 pKa = 11.84AATTTTAAAPTTTAAAPTTTAAAPTTTGAQTNVAIAVSTTPTTAPAAVPTTTARR2023 pKa = 11.84VTTTVPTTPTVQAVPTTPTVQAVPTTPTVQAAPTTVRR2060 pKa = 11.84AATTTAAAPTTAPQAATTTQRR2081 pKa = 11.84AATTTAPSTGSSGSTGSSGSGASCGGFFNLIPCIFPLTCVNGRR2124 pKa = 11.84CQLLGVIGG2132 pKa = 3.74
Molecular weight: 201.67 kDa
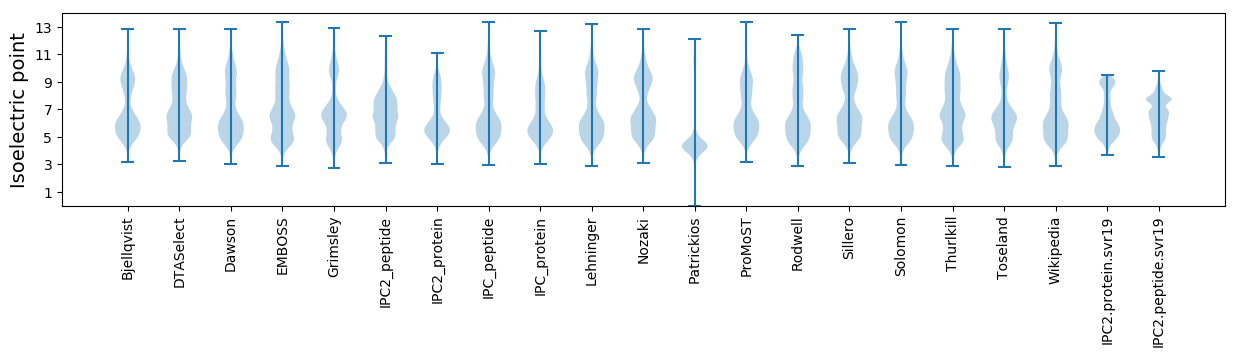
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G2XET4|G2XET4_VERDV Multiprotein-bridging factor 1 OS=Verticillium dahliae (strain VdLs.17 / ATCC MYA-4575 / FGSC 10137) OX=498257 GN=VDAG_08669 PE=3 SV=1
MM1 pKa = 7.94PILRR5 pKa = 11.84SSQPRR10 pKa = 11.84RR11 pKa = 11.84TRR13 pKa = 11.84HH14 pKa = 4.15TTTKK18 pKa = 9.78TSRR21 pKa = 11.84SNGGLFSRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84QPARR35 pKa = 11.84TTHH38 pKa = 5.14TTTTTTTTHH47 pKa = 6.21RR48 pKa = 11.84QHH50 pKa = 7.12AGATAPRR57 pKa = 11.84GGFFSRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84GPVVHH70 pKa = 6.33HH71 pKa = 5.68QQRR74 pKa = 11.84KK75 pKa = 8.1PSMGDD80 pKa = 3.22KK81 pKa = 10.49ISGAMLRR88 pKa = 11.84LKK90 pKa = 10.8GSLTRR95 pKa = 11.84RR96 pKa = 11.84PGQKK100 pKa = 9.9AAGTRR105 pKa = 11.84RR106 pKa = 11.84MNGTDD111 pKa = 2.82GRR113 pKa = 11.84GSHH116 pKa = 5.61RR117 pKa = 11.84TRR119 pKa = 11.84RR120 pKa = 11.84WW121 pKa = 2.82
MM1 pKa = 7.94PILRR5 pKa = 11.84SSQPRR10 pKa = 11.84RR11 pKa = 11.84TRR13 pKa = 11.84HH14 pKa = 4.15TTTKK18 pKa = 9.78TSRR21 pKa = 11.84SNGGLFSRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84QPARR35 pKa = 11.84TTHH38 pKa = 5.14TTTTTTTTHH47 pKa = 6.21RR48 pKa = 11.84QHH50 pKa = 7.12AGATAPRR57 pKa = 11.84GGFFSRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84GPVVHH70 pKa = 6.33HH71 pKa = 5.68QQRR74 pKa = 11.84KK75 pKa = 8.1PSMGDD80 pKa = 3.22KK81 pKa = 10.49ISGAMLRR88 pKa = 11.84LKK90 pKa = 10.8GSLTRR95 pKa = 11.84RR96 pKa = 11.84PGQKK100 pKa = 9.9AAGTRR105 pKa = 11.84RR106 pKa = 11.84MNGTDD111 pKa = 2.82GRR113 pKa = 11.84GSHH116 pKa = 5.61RR117 pKa = 11.84TRR119 pKa = 11.84RR120 pKa = 11.84WW121 pKa = 2.82
Molecular weight: 13.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4983248 |
29 |
7054 |
473.2 |
52.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.716 ± 0.024 | 1.215 ± 0.013 |
5.91 ± 0.02 | 5.95 ± 0.027 |
3.565 ± 0.015 | 7.196 ± 0.027 |
2.442 ± 0.011 | 4.412 ± 0.014 |
4.505 ± 0.024 | 8.706 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.174 ± 0.01 | 3.4 ± 0.013 |
6.286 ± 0.025 | 3.997 ± 0.017 |
6.396 ± 0.022 | 7.892 ± 0.025 |
5.984 ± 0.018 | 6.263 ± 0.017 |
1.455 ± 0.009 | 2.536 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
