
Xanthomonas phage phiLf (Bacteriophage phi-Lf)
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; unclassified Inoviridae
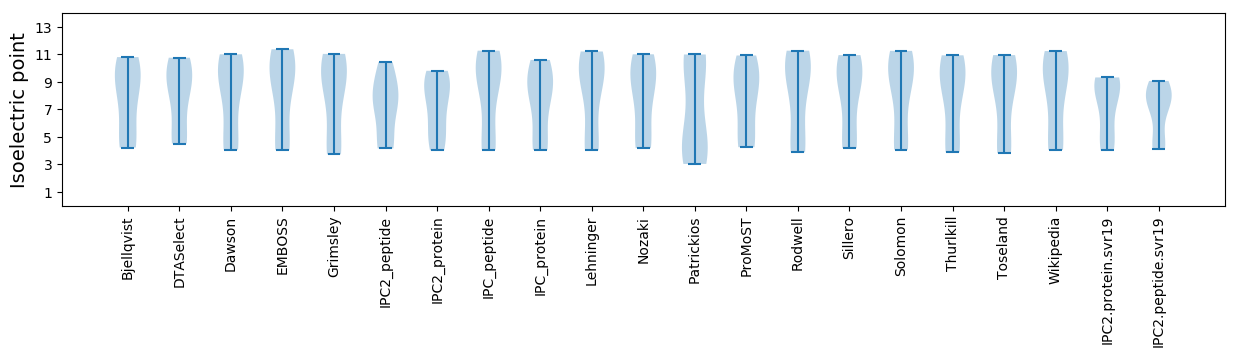
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
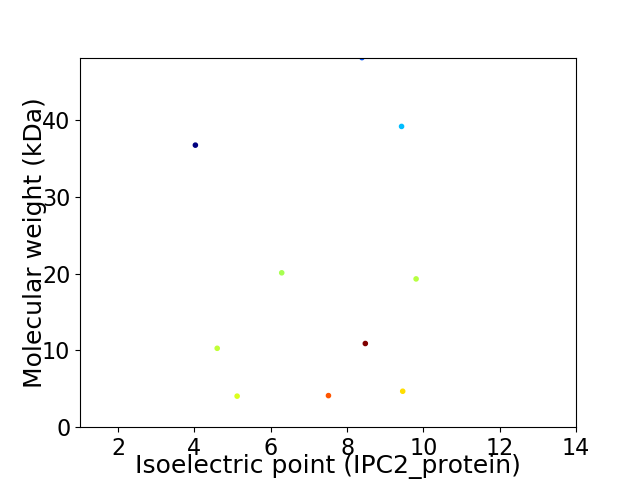
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q38617|REP_BPPHL Replication-associated protein G2P OS=Xanthomonas phage phiLf OX=28365 GN=II PE=3 SV=1
MM1 pKa = 7.39SPSLFLGWGDD11 pKa = 4.17DD12 pKa = 3.8YY13 pKa = 11.65FVVLWIHH20 pKa = 5.71GRR22 pKa = 11.84AVRR25 pKa = 11.84LGRR28 pKa = 11.84RR29 pKa = 11.84QGVGRR34 pKa = 11.84VIRR37 pKa = 11.84VSLAMLILLALTFSPIVHH55 pKa = 6.21ATCVQTEE62 pKa = 4.21PSTSANNGSWACSDD76 pKa = 3.37QGEE79 pKa = 4.47AFAKK83 pKa = 9.55ASSMGVPADD92 pKa = 4.24LSVCRR97 pKa = 11.84MKK99 pKa = 10.71SIRR102 pKa = 11.84AVSSGPGVFSQRR114 pKa = 11.84MTYY117 pKa = 10.29PGDD120 pKa = 3.51TCGIGYY126 pKa = 9.9DD127 pKa = 4.05LDD129 pKa = 4.73IGTGNATYY137 pKa = 10.39PDD139 pKa = 3.88TATCAKK145 pKa = 10.15RR146 pKa = 11.84PSQSGWTNPTAPTPSDD162 pKa = 3.5VCNDD166 pKa = 2.92GCYY169 pKa = 8.73YY170 pKa = 10.45TYY172 pKa = 11.14AVDD175 pKa = 3.65AGGPKK180 pKa = 10.35GYY182 pKa = 9.08TYY184 pKa = 10.92VPSGATCTTDD194 pKa = 4.31DD195 pKa = 4.42AAPPIDD201 pKa = 5.37DD202 pKa = 5.32GGDD205 pKa = 3.45GDD207 pKa = 5.9DD208 pKa = 5.68DD209 pKa = 4.77GGGDD213 pKa = 3.56GGGDD217 pKa = 3.3GGGDD221 pKa = 3.3GGGDD225 pKa = 3.3GGGDD229 pKa = 3.3GGGDD233 pKa = 3.3GGGDD237 pKa = 3.3GGGDD241 pKa = 3.3GGGDD245 pKa = 3.25GGGDD249 pKa = 3.53GDD251 pKa = 5.0GGGDD255 pKa = 3.82GDD257 pKa = 5.14GDD259 pKa = 4.06GDD261 pKa = 4.01GDD263 pKa = 4.09GDD265 pKa = 4.05GEE267 pKa = 4.39EE268 pKa = 4.66GGEE271 pKa = 4.07GAPMSEE277 pKa = 4.89LYY279 pKa = 10.33KK280 pKa = 10.79KK281 pKa = 10.4SGKK284 pKa = 7.4TVEE287 pKa = 4.37SVLSKK292 pKa = 10.98FNTQVRR298 pKa = 11.84GTPMVAGIGDD308 pKa = 4.1FMKK311 pKa = 10.52VPSGGSCPVFSLGASKK327 pKa = 9.08WWDD330 pKa = 2.73AMTINFHH337 pKa = 6.92CGGDD341 pKa = 3.5FLAFLRR347 pKa = 11.84AAGWVILAIAAYY359 pKa = 9.15AAIRR363 pKa = 11.84IAVTT367 pKa = 3.32
MM1 pKa = 7.39SPSLFLGWGDD11 pKa = 4.17DD12 pKa = 3.8YY13 pKa = 11.65FVVLWIHH20 pKa = 5.71GRR22 pKa = 11.84AVRR25 pKa = 11.84LGRR28 pKa = 11.84RR29 pKa = 11.84QGVGRR34 pKa = 11.84VIRR37 pKa = 11.84VSLAMLILLALTFSPIVHH55 pKa = 6.21ATCVQTEE62 pKa = 4.21PSTSANNGSWACSDD76 pKa = 3.37QGEE79 pKa = 4.47AFAKK83 pKa = 9.55ASSMGVPADD92 pKa = 4.24LSVCRR97 pKa = 11.84MKK99 pKa = 10.71SIRR102 pKa = 11.84AVSSGPGVFSQRR114 pKa = 11.84MTYY117 pKa = 10.29PGDD120 pKa = 3.51TCGIGYY126 pKa = 9.9DD127 pKa = 4.05LDD129 pKa = 4.73IGTGNATYY137 pKa = 10.39PDD139 pKa = 3.88TATCAKK145 pKa = 10.15RR146 pKa = 11.84PSQSGWTNPTAPTPSDD162 pKa = 3.5VCNDD166 pKa = 2.92GCYY169 pKa = 8.73YY170 pKa = 10.45TYY172 pKa = 11.14AVDD175 pKa = 3.65AGGPKK180 pKa = 10.35GYY182 pKa = 9.08TYY184 pKa = 10.92VPSGATCTTDD194 pKa = 4.31DD195 pKa = 4.42AAPPIDD201 pKa = 5.37DD202 pKa = 5.32GGDD205 pKa = 3.45GDD207 pKa = 5.9DD208 pKa = 5.68DD209 pKa = 4.77GGGDD213 pKa = 3.56GGGDD217 pKa = 3.3GGGDD221 pKa = 3.3GGGDD225 pKa = 3.3GGGDD229 pKa = 3.3GGGDD233 pKa = 3.3GGGDD237 pKa = 3.3GGGDD241 pKa = 3.3GGGDD245 pKa = 3.25GGGDD249 pKa = 3.53GDD251 pKa = 5.0GGGDD255 pKa = 3.82GDD257 pKa = 5.14GDD259 pKa = 4.06GDD261 pKa = 4.01GDD263 pKa = 4.09GDD265 pKa = 4.05GEE267 pKa = 4.39EE268 pKa = 4.66GGEE271 pKa = 4.07GAPMSEE277 pKa = 4.89LYY279 pKa = 10.33KK280 pKa = 10.79KK281 pKa = 10.4SGKK284 pKa = 7.4TVEE287 pKa = 4.37SVLSKK292 pKa = 10.98FNTQVRR298 pKa = 11.84GTPMVAGIGDD308 pKa = 4.1FMKK311 pKa = 10.52VPSGGSCPVFSLGASKK327 pKa = 9.08WWDD330 pKa = 2.73AMTINFHH337 pKa = 6.92CGGDD341 pKa = 3.5FLAFLRR347 pKa = 11.84AAGWVILAIAAYY359 pKa = 9.15AAIRR363 pKa = 11.84IAVTT367 pKa = 3.32
Molecular weight: 36.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q38617-2|REP-2_BPPHL Isoform of Q38617 Isoform G10P of Replication-associated protein G2P OS=Xanthomonas phage phiLf OX=28365 GN=II PE=4 SV=1
MM1 pKa = 7.78RR2 pKa = 11.84GHH4 pKa = 7.07FNRR7 pKa = 11.84LKK9 pKa = 10.44SGRR12 pKa = 11.84ARR14 pKa = 11.84WNRR17 pKa = 11.84EE18 pKa = 3.23SFRR21 pKa = 11.84YY22 pKa = 9.38VWVGEE27 pKa = 3.78LTQRR31 pKa = 11.84FRR33 pKa = 11.84PHH35 pKa = 4.67YY36 pKa = 10.3HH37 pKa = 5.23VMLWVPQGMFFGKK50 pKa = 9.92VDD52 pKa = 3.46QRR54 pKa = 11.84GWWPHH59 pKa = 5.12GSSQIEE65 pKa = 4.12KK66 pKa = 10.46ARR68 pKa = 11.84NCVGYY73 pKa = 9.54LAKK76 pKa = 10.57YY77 pKa = 9.55ASKK80 pKa = 9.13FTAITAAAFPKK91 pKa = 10.2GFRR94 pKa = 11.84THH96 pKa = 6.56GCGGLNTEE104 pKa = 4.92SKK106 pKa = 10.88RR107 pKa = 11.84EE108 pKa = 3.99LRR110 pKa = 11.84WWKK113 pKa = 10.13APKK116 pKa = 9.76DD117 pKa = 3.52ARR119 pKa = 11.84EE120 pKa = 4.1ALGGEE125 pKa = 3.86ADD127 pKa = 3.2IRR129 pKa = 11.84KK130 pKa = 9.88AKK132 pKa = 10.54GGWFDD137 pKa = 3.86RR138 pKa = 11.84LTGEE142 pKa = 4.94FWPSPWKK149 pKa = 9.18VTFIFGRR156 pKa = 11.84TFAWKK161 pKa = 10.04VVQLL165 pKa = 4.0
MM1 pKa = 7.78RR2 pKa = 11.84GHH4 pKa = 7.07FNRR7 pKa = 11.84LKK9 pKa = 10.44SGRR12 pKa = 11.84ARR14 pKa = 11.84WNRR17 pKa = 11.84EE18 pKa = 3.23SFRR21 pKa = 11.84YY22 pKa = 9.38VWVGEE27 pKa = 3.78LTQRR31 pKa = 11.84FRR33 pKa = 11.84PHH35 pKa = 4.67YY36 pKa = 10.3HH37 pKa = 5.23VMLWVPQGMFFGKK50 pKa = 9.92VDD52 pKa = 3.46QRR54 pKa = 11.84GWWPHH59 pKa = 5.12GSSQIEE65 pKa = 4.12KK66 pKa = 10.46ARR68 pKa = 11.84NCVGYY73 pKa = 9.54LAKK76 pKa = 10.57YY77 pKa = 9.55ASKK80 pKa = 9.13FTAITAAAFPKK91 pKa = 10.2GFRR94 pKa = 11.84THH96 pKa = 6.56GCGGLNTEE104 pKa = 4.92SKK106 pKa = 10.88RR107 pKa = 11.84EE108 pKa = 3.99LRR110 pKa = 11.84WWKK113 pKa = 10.13APKK116 pKa = 9.76DD117 pKa = 3.52ARR119 pKa = 11.84EE120 pKa = 4.1ALGGEE125 pKa = 3.86ADD127 pKa = 3.2IRR129 pKa = 11.84KK130 pKa = 9.88AKK132 pKa = 10.54GGWFDD137 pKa = 3.86RR138 pKa = 11.84LTGEE142 pKa = 4.94FWPSPWKK149 pKa = 9.18VTFIFGRR156 pKa = 11.84TFAWKK161 pKa = 10.04VVQLL165 pKa = 4.0
Molecular weight: 19.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1825 |
38 |
440 |
182.5 |
19.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.74 ± 0.924 | 1.589 ± 0.488 |
5.973 ± 1.152 | 3.616 ± 0.562 |
4.548 ± 0.806 | 12.164 ± 2.123 |
1.534 ± 0.33 | 3.945 ± 0.464 |
4.822 ± 0.749 | 6.411 ± 0.797 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.63 ± 0.482 | 2.466 ± 0.422 |
5.26 ± 0.543 | 3.562 ± 0.571 |
6.63 ± 1.078 | 5.973 ± 0.533 |
6.027 ± 0.436 | 6.74 ± 0.46 |
2.74 ± 0.673 | 2.63 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
