
Martelella endophytica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Martelella
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
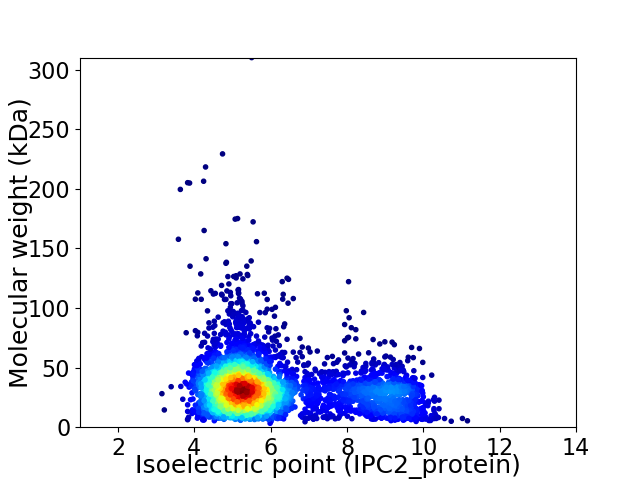
Virtual 2D-PAGE plot for 4020 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D5LKK0|A0A0D5LKK0_9RHIZ Transcriptional regulator OS=Martelella endophytica OX=1486262 GN=TM49_00245 PE=4 SV=1
MM1 pKa = 8.23DD2 pKa = 4.12SRR4 pKa = 11.84KK5 pKa = 9.92FSDD8 pKa = 3.36SNAVFFKK15 pKa = 10.79RR16 pKa = 11.84KK17 pKa = 9.57GSLITQTALSGVFAIGAVMLASSSSFSADD46 pKa = 3.3RR47 pKa = 11.84SWDD50 pKa = 3.61LFPTNGNFVVGSNWSGNTAPGAADD74 pKa = 3.47NANIGASFNSPWLDD88 pKa = 3.03QTYY91 pKa = 10.09SVQSVSVSGGGSFTIRR107 pKa = 11.84NGGNLTADD115 pKa = 3.31TMTVGGVLSSITIASGGAFNGQVSATGGTFTNSGTVNGAASISGGIATNSGSVTGMVTNSSIGLVLVPGFTNASGGSIGGLTNSGIAVNEE205 pKa = 3.96NGASIGTLSNTNGTFSNSGTVTGTTSISGGVVTNNSDD242 pKa = 3.71GVFEE246 pKa = 5.15GATSMTGGITTNNGTINGATTVSGALTSFTNNGTINNTLGITSGIVTNNGSVTGMVTNSSIGIIVAPGFTNASGGSIGGLANSGIAVNEE335 pKa = 3.96NGASIGTLSNTNGTFSNSGTVTGTTSISGGVVTNNSDD372 pKa = 3.71GVFEE376 pKa = 5.15GATSMTGGITTNNGTINGATTVSGALTSFTNNGTINNTLGITSGIVTNNGSVTGMVTNSSIGIVVAPGFTNAGGGSIGGLSNSGIAVNEE465 pKa = 3.96NGASIGTLSNTNGTFSNSGTVTGTTSISGGVVTNNSDD502 pKa = 3.71GVFEE506 pKa = 5.15GATSMTGGILTNNGSIDD523 pKa = 3.58NTLGITGGVATNNGTVAGMVTNSSFGLVLVPGFTNASGGSIGGLTNSGIAVNEE576 pKa = 3.92NGGTIGTLSNTGGVFTNSGTVTGTTTVSGGVVTNTSDD613 pKa = 3.8GVFEE617 pKa = 5.16GATSMTGGITTNNGTINGATTVSGVATTFTNTGTIDD653 pKa = 3.33NTLGVTGGIVTNTGTVTGATGVSGGLLTNTGTIDD687 pKa = 3.39NTLGVAGGVVINTGTVTGATGVSAGLLTNTGTIDD721 pKa = 3.36NTLGITGGVVVNAGTVTGAAGISGGLLTNTGTLEE755 pKa = 3.98STLDD759 pKa = 3.47VLGGFVANEE768 pKa = 3.77AGGDD772 pKa = 3.89VVGLTTIYY780 pKa = 11.04DD781 pKa = 3.69GDD783 pKa = 3.9VTNGLISAANFGTVINYY800 pKa = 7.49STGDD804 pKa = 3.22INSNGFTNGSFGRR817 pKa = 11.84TEE819 pKa = 3.68ALVNHH824 pKa = 6.87GSSVNLIGGSIGTATNNAGTFTNSGSIDD852 pKa = 3.31GALVVNGGTVDD863 pKa = 3.4NNGNGVLNNGTVGGLTTISNDD884 pKa = 3.22GLVTNSGTLADD895 pKa = 4.26VVNGSYY901 pKa = 10.73AGGSGSGFVNQAGGVAGALTNSGGATNEE929 pKa = 3.96AGGTLASVLNTAGVFTNAGTVTGTLDD955 pKa = 3.47VTGGSADD962 pKa = 3.52NTGSVAGLTTISNDD976 pKa = 3.14GVATNSGTLADD987 pKa = 3.9VVNGSFAGGSGFGFVNQSGGVAGALTNSGGATNMAGATLASVTNDD1032 pKa = 2.51AGYY1035 pKa = 7.77FTNAGTVSGSVAVNAGTAEE1054 pKa = 4.01NTGLIGGAVDD1064 pKa = 3.2IADD1067 pKa = 4.22NGTMTNSGQIDD1078 pKa = 4.38GLTTNAGLFATTGTLNGGLVNSGIASIRR1106 pKa = 11.84GTINGAIEE1114 pKa = 4.11NSSLLDD1120 pKa = 3.47DD1121 pKa = 4.32TFSLTGDD1128 pKa = 3.62LVGNGTLSNQGTFTAWGGYY1147 pKa = 7.06TISGIDD1153 pKa = 3.36IDD1155 pKa = 5.34NSGTLNASNYY1165 pKa = 8.29KK1166 pKa = 9.52TDD1168 pKa = 3.21TGALKK1173 pKa = 9.33TAGTSATTLTVDD1185 pKa = 3.67SDD1187 pKa = 3.38ITRR1190 pKa = 11.84GGRR1193 pKa = 11.84IALGTYY1199 pKa = 10.75ADD1201 pKa = 3.88TTTDD1205 pKa = 3.57QIQVTGDD1212 pKa = 3.48FSGGDD1217 pKa = 3.65YY1218 pKa = 11.35DD1219 pKa = 4.59LALYY1223 pKa = 9.24GLNYY1227 pKa = 10.22LDD1229 pKa = 3.91TPEE1232 pKa = 4.25EE1233 pKa = 4.08RR1234 pKa = 11.84TLITIDD1240 pKa = 3.38GTYY1243 pKa = 9.57SASVGDD1249 pKa = 4.31VYY1251 pKa = 11.67GLGNTANPLIVNSVRR1266 pKa = 11.84EE1267 pKa = 4.16TANSVILTTALDD1279 pKa = 3.6MTPISGTLGGIYY1291 pKa = 8.91ATQAFTGITTNRR1303 pKa = 11.84FDD1305 pKa = 4.06DD1306 pKa = 4.28SYY1308 pKa = 10.73VTAVDD1313 pKa = 4.16GAGKK1317 pKa = 10.11GCSPGTYY1324 pKa = 8.74TRR1326 pKa = 11.84MNGGSLNTASTASSAGSVSAKK1347 pKa = 8.98TDD1349 pKa = 3.04SDD1351 pKa = 3.42ISFGGVEE1358 pKa = 4.25LGLDD1362 pKa = 3.69YY1363 pKa = 11.71GCFDD1367 pKa = 4.15LGKK1370 pKa = 10.65AGGQVNFGLLGGLNSGSVSSNEE1392 pKa = 3.77TVAGIGYY1399 pKa = 9.46SGQSTFDD1406 pKa = 3.04QQYY1409 pKa = 7.79FGGYY1413 pKa = 9.92ASYY1416 pKa = 10.9LAGPFVAQLRR1426 pKa = 11.84TTYY1429 pKa = 10.79GRR1431 pKa = 11.84TAYY1434 pKa = 10.39DD1435 pKa = 3.34IDD1437 pKa = 3.68QMRR1440 pKa = 11.84TASGLSTSVFDD1451 pKa = 5.28QDD1453 pKa = 5.19GFDD1456 pKa = 3.83ANQYY1460 pKa = 6.84TASGYY1465 pKa = 9.92LAYY1468 pKa = 9.99GFNMDD1473 pKa = 4.94RR1474 pKa = 11.84FTLLPSIGLSYY1485 pKa = 11.08SNSVFNDD1492 pKa = 3.43DD1493 pKa = 4.11LQVASGSPAQISFDD1507 pKa = 4.13DD1508 pKa = 3.91MEE1510 pKa = 4.68TLVGSIGLAASASFTLDD1527 pKa = 3.46DD1528 pKa = 4.18NRR1530 pKa = 11.84GTIQPFLAGTYY1541 pKa = 9.82YY1542 pKa = 11.34NNFADD1547 pKa = 5.72DD1548 pKa = 3.41PTAYY1552 pKa = 9.51YY1553 pKa = 10.45VSSAGIRR1560 pKa = 11.84TPINTSTVDD1569 pKa = 3.17NYY1571 pKa = 11.75GEE1573 pKa = 3.9ISVGFDD1579 pKa = 3.2YY1580 pKa = 9.06LTPPTSVLRR1589 pKa = 11.84QVSGQVRR1596 pKa = 11.84ADD1598 pKa = 3.7FPVGKK1603 pKa = 9.41DD1604 pKa = 3.23VSGAQVSLSMRR1615 pKa = 11.84AQFF1618 pKa = 4.51
MM1 pKa = 8.23DD2 pKa = 4.12SRR4 pKa = 11.84KK5 pKa = 9.92FSDD8 pKa = 3.36SNAVFFKK15 pKa = 10.79RR16 pKa = 11.84KK17 pKa = 9.57GSLITQTALSGVFAIGAVMLASSSSFSADD46 pKa = 3.3RR47 pKa = 11.84SWDD50 pKa = 3.61LFPTNGNFVVGSNWSGNTAPGAADD74 pKa = 3.47NANIGASFNSPWLDD88 pKa = 3.03QTYY91 pKa = 10.09SVQSVSVSGGGSFTIRR107 pKa = 11.84NGGNLTADD115 pKa = 3.31TMTVGGVLSSITIASGGAFNGQVSATGGTFTNSGTVNGAASISGGIATNSGSVTGMVTNSSIGLVLVPGFTNASGGSIGGLTNSGIAVNEE205 pKa = 3.96NGASIGTLSNTNGTFSNSGTVTGTTSISGGVVTNNSDD242 pKa = 3.71GVFEE246 pKa = 5.15GATSMTGGITTNNGTINGATTVSGALTSFTNNGTINNTLGITSGIVTNNGSVTGMVTNSSIGIIVAPGFTNASGGSIGGLANSGIAVNEE335 pKa = 3.96NGASIGTLSNTNGTFSNSGTVTGTTSISGGVVTNNSDD372 pKa = 3.71GVFEE376 pKa = 5.15GATSMTGGITTNNGTINGATTVSGALTSFTNNGTINNTLGITSGIVTNNGSVTGMVTNSSIGIVVAPGFTNAGGGSIGGLSNSGIAVNEE465 pKa = 3.96NGASIGTLSNTNGTFSNSGTVTGTTSISGGVVTNNSDD502 pKa = 3.71GVFEE506 pKa = 5.15GATSMTGGILTNNGSIDD523 pKa = 3.58NTLGITGGVATNNGTVAGMVTNSSFGLVLVPGFTNASGGSIGGLTNSGIAVNEE576 pKa = 3.92NGGTIGTLSNTGGVFTNSGTVTGTTTVSGGVVTNTSDD613 pKa = 3.8GVFEE617 pKa = 5.16GATSMTGGITTNNGTINGATTVSGVATTFTNTGTIDD653 pKa = 3.33NTLGVTGGIVTNTGTVTGATGVSGGLLTNTGTIDD687 pKa = 3.39NTLGVAGGVVINTGTVTGATGVSAGLLTNTGTIDD721 pKa = 3.36NTLGITGGVVVNAGTVTGAAGISGGLLTNTGTLEE755 pKa = 3.98STLDD759 pKa = 3.47VLGGFVANEE768 pKa = 3.77AGGDD772 pKa = 3.89VVGLTTIYY780 pKa = 11.04DD781 pKa = 3.69GDD783 pKa = 3.9VTNGLISAANFGTVINYY800 pKa = 7.49STGDD804 pKa = 3.22INSNGFTNGSFGRR817 pKa = 11.84TEE819 pKa = 3.68ALVNHH824 pKa = 6.87GSSVNLIGGSIGTATNNAGTFTNSGSIDD852 pKa = 3.31GALVVNGGTVDD863 pKa = 3.4NNGNGVLNNGTVGGLTTISNDD884 pKa = 3.22GLVTNSGTLADD895 pKa = 4.26VVNGSYY901 pKa = 10.73AGGSGSGFVNQAGGVAGALTNSGGATNEE929 pKa = 3.96AGGTLASVLNTAGVFTNAGTVTGTLDD955 pKa = 3.47VTGGSADD962 pKa = 3.52NTGSVAGLTTISNDD976 pKa = 3.14GVATNSGTLADD987 pKa = 3.9VVNGSFAGGSGFGFVNQSGGVAGALTNSGGATNMAGATLASVTNDD1032 pKa = 2.51AGYY1035 pKa = 7.77FTNAGTVSGSVAVNAGTAEE1054 pKa = 4.01NTGLIGGAVDD1064 pKa = 3.2IADD1067 pKa = 4.22NGTMTNSGQIDD1078 pKa = 4.38GLTTNAGLFATTGTLNGGLVNSGIASIRR1106 pKa = 11.84GTINGAIEE1114 pKa = 4.11NSSLLDD1120 pKa = 3.47DD1121 pKa = 4.32TFSLTGDD1128 pKa = 3.62LVGNGTLSNQGTFTAWGGYY1147 pKa = 7.06TISGIDD1153 pKa = 3.36IDD1155 pKa = 5.34NSGTLNASNYY1165 pKa = 8.29KK1166 pKa = 9.52TDD1168 pKa = 3.21TGALKK1173 pKa = 9.33TAGTSATTLTVDD1185 pKa = 3.67SDD1187 pKa = 3.38ITRR1190 pKa = 11.84GGRR1193 pKa = 11.84IALGTYY1199 pKa = 10.75ADD1201 pKa = 3.88TTTDD1205 pKa = 3.57QIQVTGDD1212 pKa = 3.48FSGGDD1217 pKa = 3.65YY1218 pKa = 11.35DD1219 pKa = 4.59LALYY1223 pKa = 9.24GLNYY1227 pKa = 10.22LDD1229 pKa = 3.91TPEE1232 pKa = 4.25EE1233 pKa = 4.08RR1234 pKa = 11.84TLITIDD1240 pKa = 3.38GTYY1243 pKa = 9.57SASVGDD1249 pKa = 4.31VYY1251 pKa = 11.67GLGNTANPLIVNSVRR1266 pKa = 11.84EE1267 pKa = 4.16TANSVILTTALDD1279 pKa = 3.6MTPISGTLGGIYY1291 pKa = 8.91ATQAFTGITTNRR1303 pKa = 11.84FDD1305 pKa = 4.06DD1306 pKa = 4.28SYY1308 pKa = 10.73VTAVDD1313 pKa = 4.16GAGKK1317 pKa = 10.11GCSPGTYY1324 pKa = 8.74TRR1326 pKa = 11.84MNGGSLNTASTASSAGSVSAKK1347 pKa = 8.98TDD1349 pKa = 3.04SDD1351 pKa = 3.42ISFGGVEE1358 pKa = 4.25LGLDD1362 pKa = 3.69YY1363 pKa = 11.71GCFDD1367 pKa = 4.15LGKK1370 pKa = 10.65AGGQVNFGLLGGLNSGSVSSNEE1392 pKa = 3.77TVAGIGYY1399 pKa = 9.46SGQSTFDD1406 pKa = 3.04QQYY1409 pKa = 7.79FGGYY1413 pKa = 9.92ASYY1416 pKa = 10.9LAGPFVAQLRR1426 pKa = 11.84TTYY1429 pKa = 10.79GRR1431 pKa = 11.84TAYY1434 pKa = 10.39DD1435 pKa = 3.34IDD1437 pKa = 3.68QMRR1440 pKa = 11.84TASGLSTSVFDD1451 pKa = 5.28QDD1453 pKa = 5.19GFDD1456 pKa = 3.83ANQYY1460 pKa = 6.84TASGYY1465 pKa = 9.92LAYY1468 pKa = 9.99GFNMDD1473 pKa = 4.94RR1474 pKa = 11.84FTLLPSIGLSYY1485 pKa = 11.08SNSVFNDD1492 pKa = 3.43DD1493 pKa = 4.11LQVASGSPAQISFDD1507 pKa = 4.13DD1508 pKa = 3.91MEE1510 pKa = 4.68TLVGSIGLAASASFTLDD1527 pKa = 3.46DD1528 pKa = 4.18NRR1530 pKa = 11.84GTIQPFLAGTYY1541 pKa = 9.82YY1542 pKa = 11.34NNFADD1547 pKa = 5.72DD1548 pKa = 3.41PTAYY1552 pKa = 9.51YY1553 pKa = 10.45VSSAGIRR1560 pKa = 11.84TPINTSTVDD1569 pKa = 3.17NYY1571 pKa = 11.75GEE1573 pKa = 3.9ISVGFDD1579 pKa = 3.2YY1580 pKa = 9.06LTPPTSVLRR1589 pKa = 11.84QVSGQVRR1596 pKa = 11.84ADD1598 pKa = 3.7FPVGKK1603 pKa = 9.41DD1604 pKa = 3.23VSGAQVSLSMRR1615 pKa = 11.84AQFF1618 pKa = 4.51
Molecular weight: 157.68 kDa
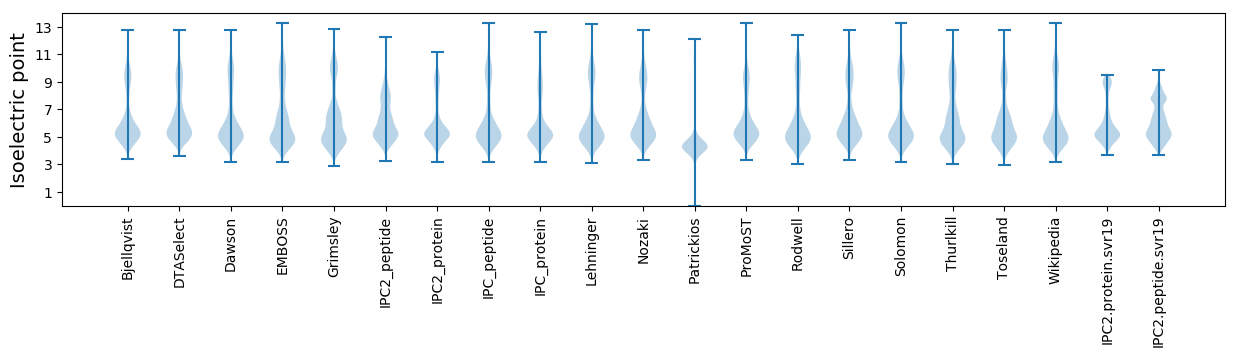
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5LPK4|A0A0D5LPK4_9RHIZ HTH marR-type domain-containing protein OS=Martelella endophytica OX=1486262 GN=TM49_11500 PE=4 SV=1
MM1 pKa = 7.58PRR3 pKa = 11.84QGPGNPGFSRR13 pKa = 11.84LTADD17 pKa = 3.72RR18 pKa = 11.84ASRR21 pKa = 11.84AASGARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84AGGRR34 pKa = 11.84HH35 pKa = 3.97SRR37 pKa = 11.84ARR39 pKa = 11.84DD40 pKa = 3.18RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84SPWRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84LPASARR56 pKa = 11.84SANRR60 pKa = 11.84ARR62 pKa = 11.84APRR65 pKa = 3.86
MM1 pKa = 7.58PRR3 pKa = 11.84QGPGNPGFSRR13 pKa = 11.84LTADD17 pKa = 3.72RR18 pKa = 11.84ASRR21 pKa = 11.84AASGARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84AGGRR34 pKa = 11.84HH35 pKa = 3.97SRR37 pKa = 11.84ARR39 pKa = 11.84DD40 pKa = 3.18RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84SPWRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84LPASARR56 pKa = 11.84SANRR60 pKa = 11.84ARR62 pKa = 11.84APRR65 pKa = 3.86
Molecular weight: 7.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1282831 |
29 |
2827 |
319.1 |
34.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.013 ± 0.05 | 0.78 ± 0.01 |
5.939 ± 0.033 | 6.047 ± 0.038 |
4.058 ± 0.027 | 8.611 ± 0.055 |
1.967 ± 0.017 | 5.64 ± 0.027 |
3.388 ± 0.031 | 9.887 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.712 ± 0.019 | 2.832 ± 0.025 |
4.943 ± 0.027 | 2.767 ± 0.022 |
6.447 ± 0.042 | 5.491 ± 0.032 |
5.443 ± 0.031 | 7.301 ± 0.034 |
1.293 ± 0.017 | 2.442 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
