
Apis mellifera associated microvirus 12
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
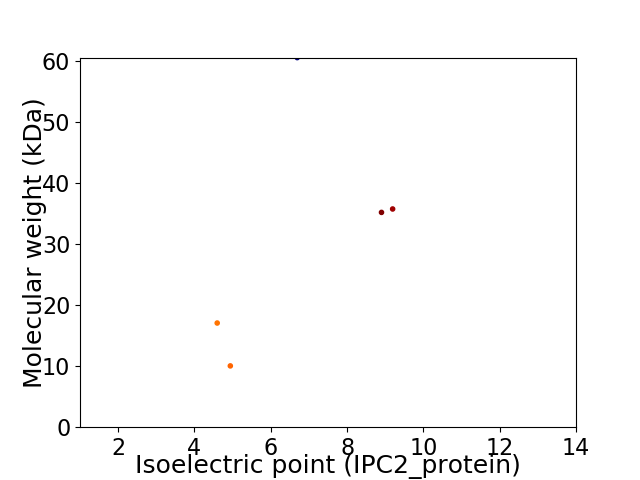
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UU62|A0A3S8UU62_9VIRU Nonstructural protein OS=Apis mellifera associated microvirus 12 OX=2494739 PE=4 SV=1
MM1 pKa = 7.64PKK3 pKa = 10.1LRR5 pKa = 11.84ARR7 pKa = 11.84RR8 pKa = 11.84GLNPLPLSPDD18 pKa = 2.94PMYY21 pKa = 10.85YY22 pKa = 9.45PQVNFGYY29 pKa = 10.75VGDD32 pKa = 3.86EE33 pKa = 4.17SDD35 pKa = 4.25RR36 pKa = 11.84VAHH39 pKa = 6.02EE40 pKa = 4.36CVPPVTAEE48 pKa = 3.72TMTRR52 pKa = 11.84QEE54 pKa = 4.89LGPATDD60 pKa = 3.21VNIIVKK66 pKa = 10.16QYY68 pKa = 10.12GASFYY73 pKa = 11.16ASGDD77 pKa = 3.19ASVPVHH83 pKa = 7.01DD84 pKa = 5.55FSLDD88 pKa = 3.33ATLLAQVRR96 pKa = 11.84RR97 pKa = 11.84EE98 pKa = 4.54ALDD101 pKa = 3.12LWSRR105 pKa = 11.84LSPRR109 pKa = 11.84QRR111 pKa = 11.84EE112 pKa = 4.33IVGSPDD118 pKa = 3.17QMVSLEE124 pKa = 4.1LQGEE128 pKa = 4.52LGAVLDD134 pKa = 4.58AVAEE138 pKa = 4.17ASSAEE143 pKa = 4.11AGGSGSPPADD153 pKa = 3.38GEE155 pKa = 4.33ATRR158 pKa = 11.84SVV160 pKa = 3.29
MM1 pKa = 7.64PKK3 pKa = 10.1LRR5 pKa = 11.84ARR7 pKa = 11.84RR8 pKa = 11.84GLNPLPLSPDD18 pKa = 2.94PMYY21 pKa = 10.85YY22 pKa = 9.45PQVNFGYY29 pKa = 10.75VGDD32 pKa = 3.86EE33 pKa = 4.17SDD35 pKa = 4.25RR36 pKa = 11.84VAHH39 pKa = 6.02EE40 pKa = 4.36CVPPVTAEE48 pKa = 3.72TMTRR52 pKa = 11.84QEE54 pKa = 4.89LGPATDD60 pKa = 3.21VNIIVKK66 pKa = 10.16QYY68 pKa = 10.12GASFYY73 pKa = 11.16ASGDD77 pKa = 3.19ASVPVHH83 pKa = 7.01DD84 pKa = 5.55FSLDD88 pKa = 3.33ATLLAQVRR96 pKa = 11.84RR97 pKa = 11.84EE98 pKa = 4.54ALDD101 pKa = 3.12LWSRR105 pKa = 11.84LSPRR109 pKa = 11.84QRR111 pKa = 11.84EE112 pKa = 4.33IVGSPDD118 pKa = 3.17QMVSLEE124 pKa = 4.1LQGEE128 pKa = 4.52LGAVLDD134 pKa = 4.58AVAEE138 pKa = 4.17ASSAEE143 pKa = 4.11AGGSGSPPADD153 pKa = 3.38GEE155 pKa = 4.33ATRR158 pKa = 11.84SVV160 pKa = 3.29
Molecular weight: 17.08 kDa
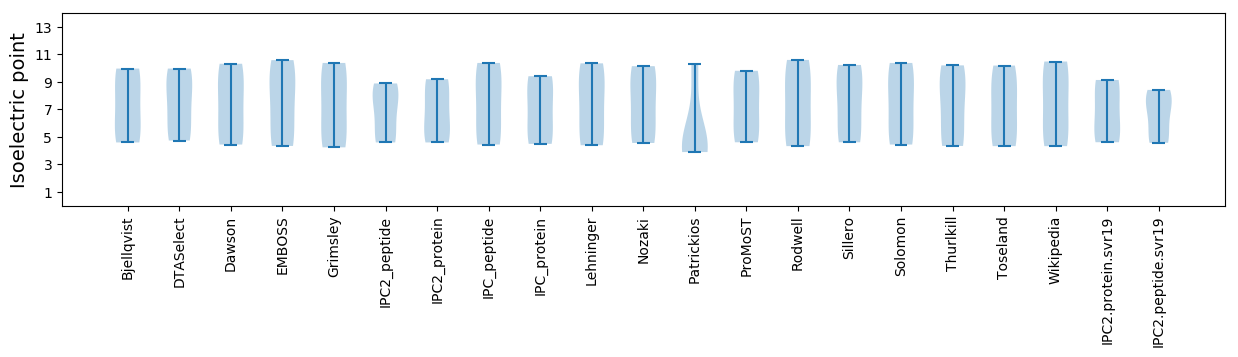
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q8U630|A0A3Q8U630_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 12 OX=2494739 PE=3 SV=1
MM1 pKa = 7.98PLPLIGGLVAGAKK14 pKa = 10.06ALGGAAMKK22 pKa = 9.76AAPWLVPSALGAVGAQRR39 pKa = 11.84TNDD42 pKa = 3.33MSRR45 pKa = 11.84DD46 pKa = 3.47LMRR49 pKa = 11.84EE50 pKa = 3.48NMAWQEE56 pKa = 3.96KK57 pKa = 8.11MANTAKK63 pKa = 10.02QRR65 pKa = 11.84AVKK68 pKa = 10.25DD69 pKa = 3.49LEE71 pKa = 4.31AAGLNPGLAYY81 pKa = 10.24GYY83 pKa = 8.34SAPMGSVSSPAMQDD97 pKa = 3.2EE98 pKa = 4.43IGAGVANAQQARR110 pKa = 11.84ATNVQLQAQRR120 pKa = 11.84ASTQKK125 pKa = 9.5IAAEE129 pKa = 3.87ARR131 pKa = 11.84IMEE134 pKa = 4.4AQAKK138 pKa = 9.31VEE140 pKa = 3.98EE141 pKa = 4.3RR142 pKa = 11.84KK143 pKa = 9.86AAEE146 pKa = 3.94FLDD149 pKa = 4.11PGVSTEE155 pKa = 4.43RR156 pKa = 11.84LNATYY161 pKa = 10.84SKK163 pKa = 10.79LRR165 pKa = 11.84AEE167 pKa = 4.46GLLGVEE173 pKa = 4.91ALRR176 pKa = 11.84QRR178 pKa = 11.84DD179 pKa = 3.56EE180 pKa = 4.16ALQTQRR186 pKa = 11.84EE187 pKa = 4.18RR188 pKa = 11.84LRR190 pKa = 11.84AIGLSNQQIEE200 pKa = 4.16QVIRR204 pKa = 11.84SGAASEE210 pKa = 4.18EE211 pKa = 4.14QTRR214 pKa = 11.84QAIAFMRR221 pKa = 11.84TMQPMEE227 pKa = 4.25VEE229 pKa = 4.27MMRR232 pKa = 11.84LRR234 pKa = 11.84QLMQKK239 pKa = 10.71LNIDD243 pKa = 3.49DD244 pKa = 3.68TRR246 pKa = 11.84YY247 pKa = 9.77GLSRR251 pKa = 11.84SRR253 pKa = 11.84AEE255 pKa = 3.55SDD257 pKa = 3.44YY258 pKa = 10.83FDD260 pKa = 4.1AVGGLGVGAEE270 pKa = 3.98RR271 pKa = 11.84LGVSRR276 pKa = 11.84LVGDD280 pKa = 4.27ALSSVTPLGRR290 pKa = 11.84MRR292 pKa = 11.84VRR294 pKa = 11.84QGAQSIKK301 pKa = 9.7EE302 pKa = 3.9SRR304 pKa = 11.84SRR306 pKa = 11.84VGAFNARR313 pKa = 11.84RR314 pKa = 11.84DD315 pKa = 3.43AEE317 pKa = 4.25AARR320 pKa = 11.84KK321 pKa = 8.84AWLEE325 pKa = 4.23SITPKK330 pKa = 10.49PP331 pKa = 3.4
MM1 pKa = 7.98PLPLIGGLVAGAKK14 pKa = 10.06ALGGAAMKK22 pKa = 9.76AAPWLVPSALGAVGAQRR39 pKa = 11.84TNDD42 pKa = 3.33MSRR45 pKa = 11.84DD46 pKa = 3.47LMRR49 pKa = 11.84EE50 pKa = 3.48NMAWQEE56 pKa = 3.96KK57 pKa = 8.11MANTAKK63 pKa = 10.02QRR65 pKa = 11.84AVKK68 pKa = 10.25DD69 pKa = 3.49LEE71 pKa = 4.31AAGLNPGLAYY81 pKa = 10.24GYY83 pKa = 8.34SAPMGSVSSPAMQDD97 pKa = 3.2EE98 pKa = 4.43IGAGVANAQQARR110 pKa = 11.84ATNVQLQAQRR120 pKa = 11.84ASTQKK125 pKa = 9.5IAAEE129 pKa = 3.87ARR131 pKa = 11.84IMEE134 pKa = 4.4AQAKK138 pKa = 9.31VEE140 pKa = 3.98EE141 pKa = 4.3RR142 pKa = 11.84KK143 pKa = 9.86AAEE146 pKa = 3.94FLDD149 pKa = 4.11PGVSTEE155 pKa = 4.43RR156 pKa = 11.84LNATYY161 pKa = 10.84SKK163 pKa = 10.79LRR165 pKa = 11.84AEE167 pKa = 4.46GLLGVEE173 pKa = 4.91ALRR176 pKa = 11.84QRR178 pKa = 11.84DD179 pKa = 3.56EE180 pKa = 4.16ALQTQRR186 pKa = 11.84EE187 pKa = 4.18RR188 pKa = 11.84LRR190 pKa = 11.84AIGLSNQQIEE200 pKa = 4.16QVIRR204 pKa = 11.84SGAASEE210 pKa = 4.18EE211 pKa = 4.14QTRR214 pKa = 11.84QAIAFMRR221 pKa = 11.84TMQPMEE227 pKa = 4.25VEE229 pKa = 4.27MMRR232 pKa = 11.84LRR234 pKa = 11.84QLMQKK239 pKa = 10.71LNIDD243 pKa = 3.49DD244 pKa = 3.68TRR246 pKa = 11.84YY247 pKa = 9.77GLSRR251 pKa = 11.84SRR253 pKa = 11.84AEE255 pKa = 3.55SDD257 pKa = 3.44YY258 pKa = 10.83FDD260 pKa = 4.1AVGGLGVGAEE270 pKa = 3.98RR271 pKa = 11.84LGVSRR276 pKa = 11.84LVGDD280 pKa = 4.27ALSSVTPLGRR290 pKa = 11.84MRR292 pKa = 11.84VRR294 pKa = 11.84QGAQSIKK301 pKa = 9.7EE302 pKa = 3.9SRR304 pKa = 11.84SRR306 pKa = 11.84VGAFNARR313 pKa = 11.84RR314 pKa = 11.84DD315 pKa = 3.43AEE317 pKa = 4.25AARR320 pKa = 11.84KK321 pKa = 8.84AWLEE325 pKa = 4.23SITPKK330 pKa = 10.49PP331 pKa = 3.4
Molecular weight: 35.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1445 |
96 |
545 |
289.0 |
31.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.588 ± 1.762 | 0.761 ± 0.444 |
5.052 ± 0.712 | 5.19 ± 0.996 |
3.253 ± 1.005 | 8.374 ± 0.428 |
2.145 ± 0.794 | 3.668 ± 0.578 |
1.938 ± 0.589 | 8.097 ± 0.393 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.699 ± 0.681 | 2.63 ± 0.803 |
6.505 ± 0.995 | 5.26 ± 0.682 |
8.789 ± 1.052 | 7.543 ± 0.837 |
5.606 ± 1.054 | 7.405 ± 0.964 |
1.799 ± 0.425 | 2.699 ± 0.36 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
