
Notothenia coriiceps (black rockcod)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia; Ctenosquamata;
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
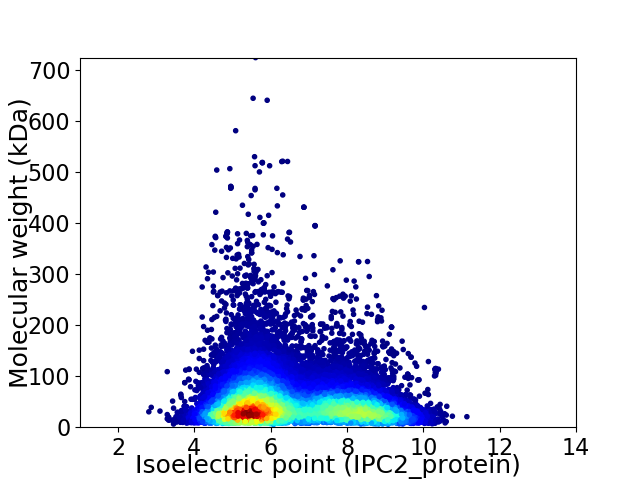
Virtual 2D-PAGE plot for 30152 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
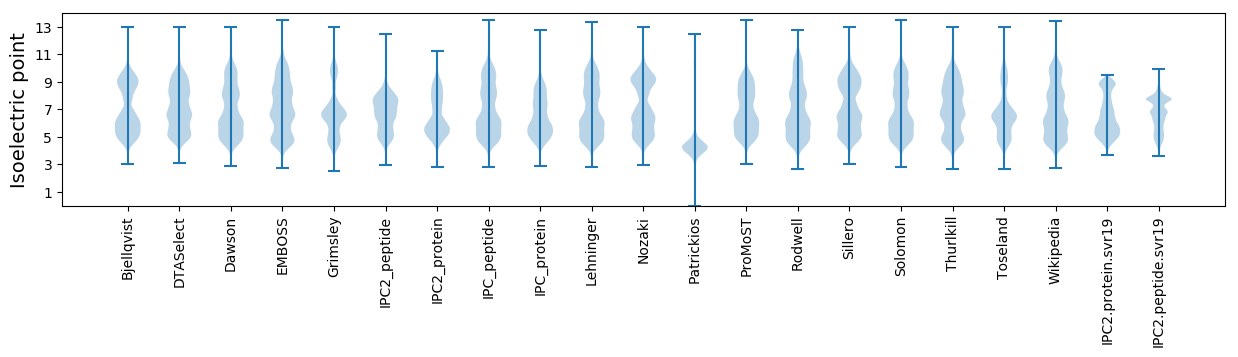
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I9N4H7|A0A6I9N4H7_9TELE trafficking protein particle complex subunit 10 OS=Notothenia coriiceps OX=8208 GN=trappc10 PE=4 SV=1
MM1 pKa = 7.51SPLFIVQQTGLVQLQSLVNSCSTSDD26 pKa = 3.57NQTSSQNNMTVDD38 pKa = 3.51MNTASSLQQYY48 pKa = 9.52RR49 pKa = 11.84SRR51 pKa = 11.84IGSSANQSPTSPVSNQGFSPGGSPQHH77 pKa = 6.1NSILGSVFADD87 pKa = 5.57FYY89 pKa = 11.38DD90 pKa = 3.5QQLPSIQASALSQQLEE106 pKa = 4.39QFNMMEE112 pKa = 4.52TPISSDD118 pKa = 2.74SLYY121 pKa = 11.11NQTSTLNYY129 pKa = 9.74SQALMMGLTGHH140 pKa = 6.96CNLQDD145 pKa = 3.89SQQLGYY151 pKa = 10.13SSHH154 pKa = 6.69GNIPNIILTVSGEE167 pKa = 4.41SPPSLPKK174 pKa = 10.74DD175 pKa = 3.36LTGCLSTDD183 pKa = 2.93VSFDD187 pKa = 3.08IDD189 pKa = 3.84TQFPLDD195 pKa = 4.35DD196 pKa = 4.83LKK198 pKa = 10.77IDD200 pKa = 4.14PLTLDD205 pKa = 4.33GLHH208 pKa = 6.4MLNDD212 pKa = 4.13PDD214 pKa = 5.58MILTDD219 pKa = 4.37PATEE223 pKa = 3.95DD224 pKa = 3.1AFRR227 pKa = 11.84LDD229 pKa = 3.51RR230 pKa = 11.84LL231 pKa = 3.74
MM1 pKa = 7.51SPLFIVQQTGLVQLQSLVNSCSTSDD26 pKa = 3.57NQTSSQNNMTVDD38 pKa = 3.51MNTASSLQQYY48 pKa = 9.52RR49 pKa = 11.84SRR51 pKa = 11.84IGSSANQSPTSPVSNQGFSPGGSPQHH77 pKa = 6.1NSILGSVFADD87 pKa = 5.57FYY89 pKa = 11.38DD90 pKa = 3.5QQLPSIQASALSQQLEE106 pKa = 4.39QFNMMEE112 pKa = 4.52TPISSDD118 pKa = 2.74SLYY121 pKa = 11.11NQTSTLNYY129 pKa = 9.74SQALMMGLTGHH140 pKa = 6.96CNLQDD145 pKa = 3.89SQQLGYY151 pKa = 10.13SSHH154 pKa = 6.69GNIPNIILTVSGEE167 pKa = 4.41SPPSLPKK174 pKa = 10.74DD175 pKa = 3.36LTGCLSTDD183 pKa = 2.93VSFDD187 pKa = 3.08IDD189 pKa = 3.84TQFPLDD195 pKa = 4.35DD196 pKa = 4.83LKK198 pKa = 10.77IDD200 pKa = 4.14PLTLDD205 pKa = 4.33GLHH208 pKa = 6.4MLNDD212 pKa = 4.13PDD214 pKa = 5.58MILTDD219 pKa = 4.37PATEE223 pKa = 3.95DD224 pKa = 3.1AFRR227 pKa = 11.84LDD229 pKa = 3.51RR230 pKa = 11.84LL231 pKa = 3.74
Molecular weight: 25.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I9PLI8|A0A6I9PLI8_9TELE Isoform of A0A6I9PMS2 uncharacterized protein LOC104961826 isoform X2 OS=Notothenia coriiceps OX=8208 GN=LOC104961826 PE=4 SV=1
MM1 pKa = 7.56SAPSSQKK8 pKa = 10.15VVLKK12 pKa = 9.94STTKK16 pKa = 9.76VSLNEE21 pKa = 4.01RR22 pKa = 11.84FTNMLKK28 pKa = 10.37NKK30 pKa = 9.57QPAAVSIRR38 pKa = 11.84ATMQQQHH45 pKa = 6.32LASARR50 pKa = 11.84NRR52 pKa = 11.84RR53 pKa = 11.84LAQQMEE59 pKa = 4.32NRR61 pKa = 11.84PSVQAALNHH70 pKa = 5.68KK71 pKa = 10.23QSLKK75 pKa = 10.22QRR77 pKa = 11.84LGKK80 pKa = 10.77SNIQARR86 pKa = 11.84LGRR89 pKa = 11.84PVGALMRR96 pKa = 11.84GGAPGSRR103 pKa = 11.84GGMRR107 pKa = 11.84GMNRR111 pKa = 11.84GGLRR115 pKa = 11.84GRR117 pKa = 11.84ARR119 pKa = 11.84GGIMRR124 pKa = 11.84GALSLRR130 pKa = 11.84GKK132 pKa = 10.08RR133 pKa = 11.84VASTGGPMRR142 pKa = 11.84GRR144 pKa = 11.84GSAGRR149 pKa = 11.84LAMRR153 pKa = 11.84RR154 pKa = 11.84AGRR157 pKa = 11.84NRR159 pKa = 11.84GGPVGRR165 pKa = 11.84GGALSRR171 pKa = 11.84GAARR175 pKa = 11.84GAARR179 pKa = 11.84GGVARR184 pKa = 11.84GG185 pKa = 3.26
MM1 pKa = 7.56SAPSSQKK8 pKa = 10.15VVLKK12 pKa = 9.94STTKK16 pKa = 9.76VSLNEE21 pKa = 4.01RR22 pKa = 11.84FTNMLKK28 pKa = 10.37NKK30 pKa = 9.57QPAAVSIRR38 pKa = 11.84ATMQQQHH45 pKa = 6.32LASARR50 pKa = 11.84NRR52 pKa = 11.84RR53 pKa = 11.84LAQQMEE59 pKa = 4.32NRR61 pKa = 11.84PSVQAALNHH70 pKa = 5.68KK71 pKa = 10.23QSLKK75 pKa = 10.22QRR77 pKa = 11.84LGKK80 pKa = 10.77SNIQARR86 pKa = 11.84LGRR89 pKa = 11.84PVGALMRR96 pKa = 11.84GGAPGSRR103 pKa = 11.84GGMRR107 pKa = 11.84GMNRR111 pKa = 11.84GGLRR115 pKa = 11.84GRR117 pKa = 11.84ARR119 pKa = 11.84GGIMRR124 pKa = 11.84GALSLRR130 pKa = 11.84GKK132 pKa = 10.08RR133 pKa = 11.84VASTGGPMRR142 pKa = 11.84GRR144 pKa = 11.84GSAGRR149 pKa = 11.84LAMRR153 pKa = 11.84RR154 pKa = 11.84AGRR157 pKa = 11.84NRR159 pKa = 11.84GGPVGRR165 pKa = 11.84GGALSRR171 pKa = 11.84GAARR175 pKa = 11.84GAARR179 pKa = 11.84GGVARR184 pKa = 11.84GG185 pKa = 3.26
Molecular weight: 19.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
14412374 |
37 |
7994 |
478.0 |
53.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.565 ± 0.013 | 2.123 ± 0.013 |
5.206 ± 0.01 | 7.08 ± 0.021 |
3.416 ± 0.01 | 6.536 ± 0.027 |
2.647 ± 0.008 | 4.178 ± 0.01 |
5.658 ± 0.016 | 9.247 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.443 ± 0.006 | 3.809 ± 0.008 |
6.035 ± 0.019 | 4.864 ± 0.015 |
5.672 ± 0.012 | 9.046 ± 0.021 |
5.645 ± 0.012 | 6.156 ± 0.012 |
1.075 ± 0.005 | 2.594 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
