
Dielma fastidiosa
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Erysipelotrichia; Erysipelotrichales; Erysipelotrichaceae; Dielma
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
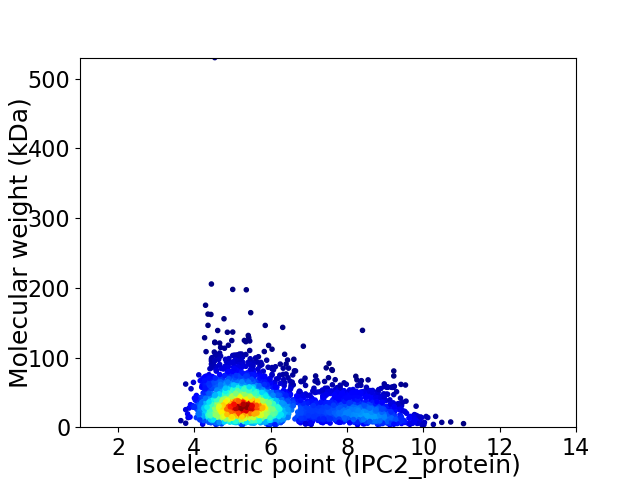
Virtual 2D-PAGE plot for 3467 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A318L5U5|A0A318L5U5_9FIRM Putative ABC transport system ATP-binding protein OS=Dielma fastidiosa OX=1034346 GN=DES51_1126 PE=4 SV=1
MM1 pKa = 6.99FAAIIGKK8 pKa = 9.8GKK10 pKa = 10.26LVLLIFCLLSSMPMHH25 pKa = 6.57SFLEE29 pKa = 4.11IEE31 pKa = 4.22EE32 pKa = 4.47WEE34 pKa = 4.27DD35 pKa = 3.02TGNEE39 pKa = 3.97VLEE42 pKa = 4.92DD43 pKa = 3.51EE44 pKa = 5.39GEE46 pKa = 4.47SADD49 pKa = 4.96HH50 pKa = 6.81EE51 pKa = 4.77SADD54 pKa = 3.99EE55 pKa = 4.46DD56 pKa = 4.21SDD58 pKa = 4.17YY59 pKa = 11.73QEE61 pKa = 5.09ADD63 pKa = 2.84QGEE66 pKa = 4.65DD67 pKa = 3.62LNEE70 pKa = 3.94ATAGDD75 pKa = 4.38ILPDD79 pKa = 3.72DD80 pKa = 4.17TASDD84 pKa = 4.13SDD86 pKa = 3.92SSSDD90 pKa = 4.1IIEE93 pKa = 4.57NDD95 pKa = 3.76PLPAEE100 pKa = 4.56PSDD103 pKa = 3.9TNAEE107 pKa = 3.92PSLPDD112 pKa = 3.39TNGVCEE118 pKa = 4.35VEE120 pKa = 4.25DD121 pKa = 3.46ALGFRR126 pKa = 11.84EE127 pKa = 4.84CLAQEE132 pKa = 3.73KK133 pKa = 10.29DD134 pKa = 3.1IRR136 pKa = 11.84LTLLSDD142 pKa = 3.68VSVNAADD149 pKa = 4.87LPNADD154 pKa = 4.49TEE156 pKa = 4.36FLSIDD161 pKa = 3.9LNGHH165 pKa = 5.41QLTIVIDD172 pKa = 3.76EE173 pKa = 5.01DD174 pKa = 5.19SNFTPLRR181 pKa = 11.84CSSFQLISEE190 pKa = 4.39SEE192 pKa = 4.19HH193 pKa = 8.7AGMLSVKK200 pKa = 9.93AASSMSAPFLAVSSYY215 pKa = 11.72AFFAGSEE222 pKa = 4.07AGIQIYY228 pKa = 9.26SQSADD233 pKa = 3.14KK234 pKa = 10.56QAYY237 pKa = 7.97TMIDD241 pKa = 4.08LIGDD245 pKa = 3.87AEE247 pKa = 4.2MNAQNSRR254 pKa = 11.84FMTNTGDD261 pKa = 3.56VLTLITRR268 pKa = 11.84EE269 pKa = 4.23GNKK272 pKa = 9.94LPQTAWIEE280 pKa = 3.87FANIDD285 pKa = 3.44QHH287 pKa = 9.42AEE289 pKa = 3.82FLNQQILYY297 pKa = 9.19YY298 pKa = 8.95YY299 pKa = 10.43LPASPFLYY307 pKa = 10.67AEE309 pKa = 3.91SDD311 pKa = 3.37KK312 pKa = 11.41YY313 pKa = 10.27QVQIEE318 pKa = 4.31WGDD321 pKa = 3.36LSYY324 pKa = 11.26SYY326 pKa = 11.35DD327 pKa = 3.68GDD329 pKa = 3.66RR330 pKa = 11.84MSWQGNDD337 pKa = 3.04NLNNHH342 pKa = 5.81IRR344 pKa = 11.84VNNYY348 pKa = 9.76SLFPVMITHH357 pKa = 7.3EE358 pKa = 4.3LQLDD362 pKa = 3.7DD363 pKa = 5.51GNIKK367 pKa = 10.68GDD369 pKa = 3.59VYY371 pKa = 10.18KK372 pKa = 10.26TAGSKK377 pKa = 10.55SDD379 pKa = 3.64GSDD382 pKa = 3.06EE383 pKa = 4.13YY384 pKa = 11.36DD385 pKa = 3.4YY386 pKa = 12.05DD387 pKa = 3.79MCEE390 pKa = 3.4LAAYY394 pKa = 9.69EE395 pKa = 4.4EE396 pKa = 4.37EE397 pKa = 5.02LIAAASNLPTQLDD410 pKa = 4.39LYY412 pKa = 10.87VVLSGNPDD420 pKa = 3.22LDD422 pKa = 3.58QYY424 pKa = 11.42KK425 pKa = 9.44HH426 pKa = 5.39QKK428 pKa = 9.98QIGSLSLTVTSMEE441 pKa = 5.09NEE443 pKa = 3.97PQNLNEE449 pKa = 4.09PAEE452 pKa = 4.39AFDD455 pKa = 3.67LTIEE459 pKa = 4.19KK460 pKa = 10.86GEE462 pKa = 4.02TLILSGEE469 pKa = 4.13HH470 pKa = 5.85EE471 pKa = 4.72MNGNIYY477 pKa = 9.73RR478 pKa = 11.84VSGGGTLCFDD488 pKa = 3.45QFQAVDD494 pKa = 3.73ALDD497 pKa = 4.11GDD499 pKa = 4.32VLAEE503 pKa = 4.12CDD505 pKa = 3.9EE506 pKa = 4.78DD507 pKa = 4.41SSLIAGSHH515 pKa = 5.42QDD517 pKa = 3.17LNAYY521 pKa = 10.04RR522 pKa = 11.84FIDD525 pKa = 3.75LLGEE529 pKa = 4.06EE530 pKa = 4.62TTAGTPAVKK539 pKa = 10.43LIYY542 pKa = 9.98RR543 pKa = 11.84KK544 pKa = 10.3EE545 pKa = 4.04EE546 pKa = 4.1ADD548 pKa = 3.7VKK550 pKa = 11.51ANDD553 pKa = 3.84TNEE556 pKa = 4.26TADD559 pKa = 3.95EE560 pKa = 4.21QPQQ563 pKa = 3.16
MM1 pKa = 6.99FAAIIGKK8 pKa = 9.8GKK10 pKa = 10.26LVLLIFCLLSSMPMHH25 pKa = 6.57SFLEE29 pKa = 4.11IEE31 pKa = 4.22EE32 pKa = 4.47WEE34 pKa = 4.27DD35 pKa = 3.02TGNEE39 pKa = 3.97VLEE42 pKa = 4.92DD43 pKa = 3.51EE44 pKa = 5.39GEE46 pKa = 4.47SADD49 pKa = 4.96HH50 pKa = 6.81EE51 pKa = 4.77SADD54 pKa = 3.99EE55 pKa = 4.46DD56 pKa = 4.21SDD58 pKa = 4.17YY59 pKa = 11.73QEE61 pKa = 5.09ADD63 pKa = 2.84QGEE66 pKa = 4.65DD67 pKa = 3.62LNEE70 pKa = 3.94ATAGDD75 pKa = 4.38ILPDD79 pKa = 3.72DD80 pKa = 4.17TASDD84 pKa = 4.13SDD86 pKa = 3.92SSSDD90 pKa = 4.1IIEE93 pKa = 4.57NDD95 pKa = 3.76PLPAEE100 pKa = 4.56PSDD103 pKa = 3.9TNAEE107 pKa = 3.92PSLPDD112 pKa = 3.39TNGVCEE118 pKa = 4.35VEE120 pKa = 4.25DD121 pKa = 3.46ALGFRR126 pKa = 11.84EE127 pKa = 4.84CLAQEE132 pKa = 3.73KK133 pKa = 10.29DD134 pKa = 3.1IRR136 pKa = 11.84LTLLSDD142 pKa = 3.68VSVNAADD149 pKa = 4.87LPNADD154 pKa = 4.49TEE156 pKa = 4.36FLSIDD161 pKa = 3.9LNGHH165 pKa = 5.41QLTIVIDD172 pKa = 3.76EE173 pKa = 5.01DD174 pKa = 5.19SNFTPLRR181 pKa = 11.84CSSFQLISEE190 pKa = 4.39SEE192 pKa = 4.19HH193 pKa = 8.7AGMLSVKK200 pKa = 9.93AASSMSAPFLAVSSYY215 pKa = 11.72AFFAGSEE222 pKa = 4.07AGIQIYY228 pKa = 9.26SQSADD233 pKa = 3.14KK234 pKa = 10.56QAYY237 pKa = 7.97TMIDD241 pKa = 4.08LIGDD245 pKa = 3.87AEE247 pKa = 4.2MNAQNSRR254 pKa = 11.84FMTNTGDD261 pKa = 3.56VLTLITRR268 pKa = 11.84EE269 pKa = 4.23GNKK272 pKa = 9.94LPQTAWIEE280 pKa = 3.87FANIDD285 pKa = 3.44QHH287 pKa = 9.42AEE289 pKa = 3.82FLNQQILYY297 pKa = 9.19YY298 pKa = 8.95YY299 pKa = 10.43LPASPFLYY307 pKa = 10.67AEE309 pKa = 3.91SDD311 pKa = 3.37KK312 pKa = 11.41YY313 pKa = 10.27QVQIEE318 pKa = 4.31WGDD321 pKa = 3.36LSYY324 pKa = 11.26SYY326 pKa = 11.35DD327 pKa = 3.68GDD329 pKa = 3.66RR330 pKa = 11.84MSWQGNDD337 pKa = 3.04NLNNHH342 pKa = 5.81IRR344 pKa = 11.84VNNYY348 pKa = 9.76SLFPVMITHH357 pKa = 7.3EE358 pKa = 4.3LQLDD362 pKa = 3.7DD363 pKa = 5.51GNIKK367 pKa = 10.68GDD369 pKa = 3.59VYY371 pKa = 10.18KK372 pKa = 10.26TAGSKK377 pKa = 10.55SDD379 pKa = 3.64GSDD382 pKa = 3.06EE383 pKa = 4.13YY384 pKa = 11.36DD385 pKa = 3.4YY386 pKa = 12.05DD387 pKa = 3.79MCEE390 pKa = 3.4LAAYY394 pKa = 9.69EE395 pKa = 4.4EE396 pKa = 4.37EE397 pKa = 5.02LIAAASNLPTQLDD410 pKa = 4.39LYY412 pKa = 10.87VVLSGNPDD420 pKa = 3.22LDD422 pKa = 3.58QYY424 pKa = 11.42KK425 pKa = 9.44HH426 pKa = 5.39QKK428 pKa = 9.98QIGSLSLTVTSMEE441 pKa = 5.09NEE443 pKa = 3.97PQNLNEE449 pKa = 4.09PAEE452 pKa = 4.39AFDD455 pKa = 3.67LTIEE459 pKa = 4.19KK460 pKa = 10.86GEE462 pKa = 4.02TLILSGEE469 pKa = 4.13HH470 pKa = 5.85EE471 pKa = 4.72MNGNIYY477 pKa = 9.73RR478 pKa = 11.84VSGGGTLCFDD488 pKa = 3.45QFQAVDD494 pKa = 3.73ALDD497 pKa = 4.11GDD499 pKa = 4.32VLAEE503 pKa = 4.12CDD505 pKa = 3.9EE506 pKa = 4.78DD507 pKa = 4.41SSLIAGSHH515 pKa = 5.42QDD517 pKa = 3.17LNAYY521 pKa = 10.04RR522 pKa = 11.84FIDD525 pKa = 3.75LLGEE529 pKa = 4.06EE530 pKa = 4.62TTAGTPAVKK539 pKa = 10.43LIYY542 pKa = 9.98RR543 pKa = 11.84KK544 pKa = 10.3EE545 pKa = 4.04EE546 pKa = 4.1ADD548 pKa = 3.7VKK550 pKa = 11.51ANDD553 pKa = 3.84TNEE556 pKa = 4.26TADD559 pKa = 3.95EE560 pKa = 4.21QPQQ563 pKa = 3.16
Molecular weight: 61.9 kDa
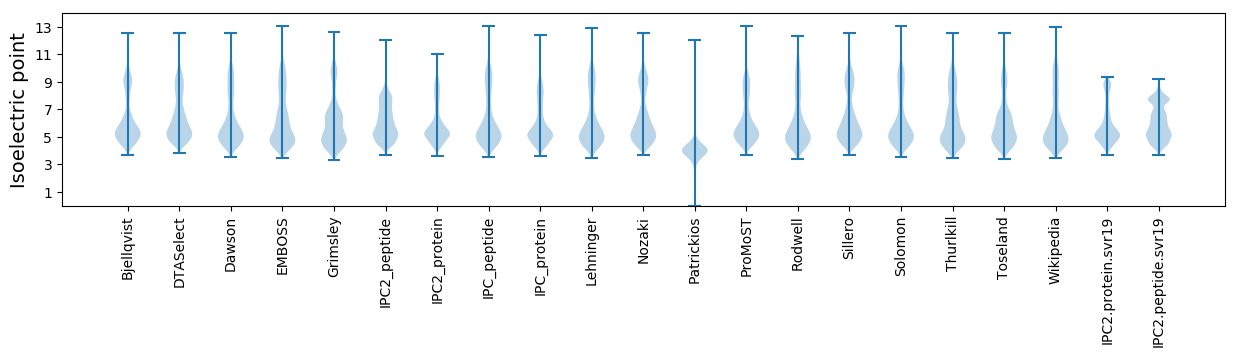
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V2FF10|A0A2V2FF10_9FIRM PTS sugar transporter subunit IIC OS=Dielma fastidiosa OX=1034346 GN=DES51_102182 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.14QPSKK9 pKa = 10.05RR10 pKa = 11.84KK11 pKa = 8.47HH12 pKa = 5.31QKK14 pKa = 6.82THH16 pKa = 4.76GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.59GRR39 pKa = 11.84KK40 pKa = 8.44VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.14QPSKK9 pKa = 10.05RR10 pKa = 11.84KK11 pKa = 8.47HH12 pKa = 5.31QKK14 pKa = 6.82THH16 pKa = 4.76GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.59GRR39 pKa = 11.84KK40 pKa = 8.44VLSAA44 pKa = 4.05
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1067974 |
27 |
4918 |
308.0 |
34.63 |
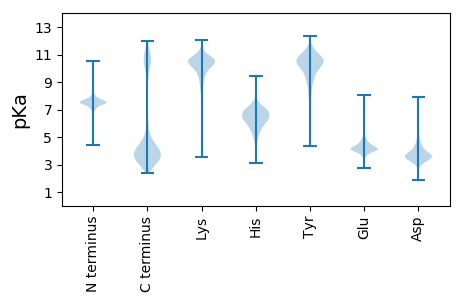
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.617 ± 0.048 | 1.563 ± 0.019 |
5.807 ± 0.038 | 6.907 ± 0.04 |
4.294 ± 0.031 | 6.256 ± 0.052 |
1.968 ± 0.023 | 8.035 ± 0.042 |
6.727 ± 0.036 | 9.966 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.127 ± 0.023 | 4.925 ± 0.032 |
3.05 ± 0.02 | 3.623 ± 0.028 |
3.663 ± 0.032 | 5.928 ± 0.034 |
5.066 ± 0.041 | 6.409 ± 0.035 |
0.856 ± 0.015 | 4.212 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
