
Notechis scutatus (mainland tiger snake)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida;
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
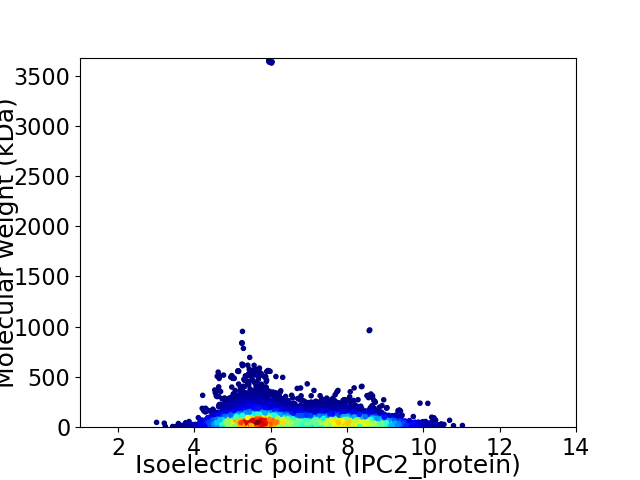
Virtual 2D-PAGE plot for 26729 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6J1W5P7|A0A6J1W5P7_9SAUR GTP-binding protein 1 OS=Notechis scutatus OX=8663 GN=GTPBP1 PE=4 SV=1
CC1 pKa = 7.44LFANPCFAGSKK12 pKa = 9.47CLNIAPGFRR21 pKa = 11.84CEE23 pKa = 4.09PCPLGYY29 pKa = 10.2KK30 pKa = 10.29GNLVTGVGADD40 pKa = 3.39YY41 pKa = 11.32AKK43 pKa = 10.6ASKK46 pKa = 10.12QICTDD51 pKa = 2.87VDD53 pKa = 3.5EE54 pKa = 5.98CNDD57 pKa = 3.93GNNGGCDD64 pKa = 3.43PNAICTNTVGSFKK77 pKa = 10.74CGPCKK82 pKa = 10.51SGFVEE87 pKa = 5.16KK88 pKa = 10.59EE89 pKa = 4.02PGSCTPQKK97 pKa = 10.57ACEE100 pKa = 4.33SPSHH104 pKa = 6.11NPCDD108 pKa = 3.86VNGYY112 pKa = 9.86CLFEE116 pKa = 4.89RR117 pKa = 11.84NGDD120 pKa = 3.75VSCSCNVGWAGNGNVCGRR138 pKa = 11.84DD139 pKa = 3.42TDD141 pKa = 3.37IDD143 pKa = 4.4GYY145 pKa = 10.04PDD147 pKa = 3.67EE148 pKa = 5.4PLPCIDD154 pKa = 4.3NNKK157 pKa = 9.47HH158 pKa = 5.93CAQDD162 pKa = 3.59NCRR165 pKa = 11.84LTPNSGQEE173 pKa = 3.95DD174 pKa = 4.26ADD176 pKa = 3.62NDD178 pKa = 5.11GIGDD182 pKa = 3.82QCDD185 pKa = 3.57DD186 pKa = 4.52DD187 pKa = 6.87ADD189 pKa = 4.09GDD191 pKa = 4.37GIKK194 pKa = 10.51NVEE197 pKa = 4.41DD198 pKa = 3.59NCRR201 pKa = 11.84LLPNKK206 pKa = 9.71DD207 pKa = 3.66QQNSDD212 pKa = 3.62PDD214 pKa = 4.13SFGDD218 pKa = 4.57ACDD221 pKa = 3.62NCPNVPNNDD230 pKa = 4.18QKK232 pKa = 11.26DD233 pKa = 3.45TDD235 pKa = 3.74QNGEE239 pKa = 3.91GDD241 pKa = 3.73ACDD244 pKa = 3.98NDD246 pKa = 3.39IDD248 pKa = 5.49GDD250 pKa = 4.41GIPNGLDD257 pKa = 3.15NCPKK261 pKa = 10.23VPNPLQTDD269 pKa = 3.4RR270 pKa = 11.84DD271 pKa = 3.67EE272 pKa = 6.42DD273 pKa = 4.4GVGDD277 pKa = 4.27ACDD280 pKa = 3.57SCPEE284 pKa = 4.33LSNPTQTDD292 pKa = 2.88VDD294 pKa = 4.03SDD296 pKa = 4.44LVGDD300 pKa = 4.12ACDD303 pKa = 3.65TNEE306 pKa = 4.67DD307 pKa = 3.81RR308 pKa = 11.84DD309 pKa = 4.1GDD311 pKa = 3.98GHH313 pKa = 7.3QDD315 pKa = 3.14TKK317 pKa = 11.44DD318 pKa = 3.42NCVEE322 pKa = 4.27IPNSSQLDD330 pKa = 3.68SDD332 pKa = 4.3NDD334 pKa = 4.23GQGDD338 pKa = 4.0DD339 pKa = 5.38CDD341 pKa = 5.28NDD343 pKa = 4.19DD344 pKa = 5.42DD345 pKa = 5.52NDD347 pKa = 5.14GIPDD351 pKa = 4.17YY352 pKa = 10.95LPPGPDD358 pKa = 3.02NCRR361 pKa = 11.84LIANPNQKK369 pKa = 10.35DD370 pKa = 3.34VDD372 pKa = 4.1GNGVGDD378 pKa = 3.67ACEE381 pKa = 3.98EE382 pKa = 4.22DD383 pKa = 3.85FDD385 pKa = 5.35NDD387 pKa = 3.74TVADD391 pKa = 5.0PIDD394 pKa = 3.65VCPEE398 pKa = 3.74SSEE401 pKa = 4.04VTLTDD406 pKa = 3.73FRR408 pKa = 11.84AYY410 pKa = 8.36QTVILDD416 pKa = 3.91PEE418 pKa = 5.2GDD420 pKa = 3.69AQIDD424 pKa = 4.13PNWVVLNQGMEE435 pKa = 4.11IVQTMNSDD443 pKa = 3.19PGLAVGYY450 pKa = 7.31TAFNGVDD457 pKa = 3.99FEE459 pKa = 4.97GTFHH463 pKa = 6.68VNTMTDD469 pKa = 2.95DD470 pKa = 4.36DD471 pKa = 4.35YY472 pKa = 12.02AGFIFAYY479 pKa = 9.53QDD481 pKa = 3.19SASFYY486 pKa = 10.36VVMWKK491 pKa = 7.78QTEE494 pKa = 3.86QTYY497 pKa = 8.13WQATPFRR504 pKa = 11.84AVAEE508 pKa = 4.33SSLQLKK514 pKa = 9.19AVKK517 pKa = 10.44SEE519 pKa = 4.18TGPGEE524 pKa = 4.19YY525 pKa = 10.12LRR527 pKa = 11.84NALWHH532 pKa = 6.17TGHH535 pKa = 6.64TPGHH539 pKa = 6.2VKK541 pKa = 10.49LLWKK545 pKa = 10.39DD546 pKa = 3.35PRR548 pKa = 11.84NVGWKK553 pKa = 10.34DD554 pKa = 2.96KK555 pKa = 10.8TSYY558 pKa = 10.16RR559 pKa = 11.84WRR561 pKa = 11.84LLHH564 pKa = 6.59RR565 pKa = 11.84PQVGYY570 pKa = 10.39IRR572 pKa = 4.61
CC1 pKa = 7.44LFANPCFAGSKK12 pKa = 9.47CLNIAPGFRR21 pKa = 11.84CEE23 pKa = 4.09PCPLGYY29 pKa = 10.2KK30 pKa = 10.29GNLVTGVGADD40 pKa = 3.39YY41 pKa = 11.32AKK43 pKa = 10.6ASKK46 pKa = 10.12QICTDD51 pKa = 2.87VDD53 pKa = 3.5EE54 pKa = 5.98CNDD57 pKa = 3.93GNNGGCDD64 pKa = 3.43PNAICTNTVGSFKK77 pKa = 10.74CGPCKK82 pKa = 10.51SGFVEE87 pKa = 5.16KK88 pKa = 10.59EE89 pKa = 4.02PGSCTPQKK97 pKa = 10.57ACEE100 pKa = 4.33SPSHH104 pKa = 6.11NPCDD108 pKa = 3.86VNGYY112 pKa = 9.86CLFEE116 pKa = 4.89RR117 pKa = 11.84NGDD120 pKa = 3.75VSCSCNVGWAGNGNVCGRR138 pKa = 11.84DD139 pKa = 3.42TDD141 pKa = 3.37IDD143 pKa = 4.4GYY145 pKa = 10.04PDD147 pKa = 3.67EE148 pKa = 5.4PLPCIDD154 pKa = 4.3NNKK157 pKa = 9.47HH158 pKa = 5.93CAQDD162 pKa = 3.59NCRR165 pKa = 11.84LTPNSGQEE173 pKa = 3.95DD174 pKa = 4.26ADD176 pKa = 3.62NDD178 pKa = 5.11GIGDD182 pKa = 3.82QCDD185 pKa = 3.57DD186 pKa = 4.52DD187 pKa = 6.87ADD189 pKa = 4.09GDD191 pKa = 4.37GIKK194 pKa = 10.51NVEE197 pKa = 4.41DD198 pKa = 3.59NCRR201 pKa = 11.84LLPNKK206 pKa = 9.71DD207 pKa = 3.66QQNSDD212 pKa = 3.62PDD214 pKa = 4.13SFGDD218 pKa = 4.57ACDD221 pKa = 3.62NCPNVPNNDD230 pKa = 4.18QKK232 pKa = 11.26DD233 pKa = 3.45TDD235 pKa = 3.74QNGEE239 pKa = 3.91GDD241 pKa = 3.73ACDD244 pKa = 3.98NDD246 pKa = 3.39IDD248 pKa = 5.49GDD250 pKa = 4.41GIPNGLDD257 pKa = 3.15NCPKK261 pKa = 10.23VPNPLQTDD269 pKa = 3.4RR270 pKa = 11.84DD271 pKa = 3.67EE272 pKa = 6.42DD273 pKa = 4.4GVGDD277 pKa = 4.27ACDD280 pKa = 3.57SCPEE284 pKa = 4.33LSNPTQTDD292 pKa = 2.88VDD294 pKa = 4.03SDD296 pKa = 4.44LVGDD300 pKa = 4.12ACDD303 pKa = 3.65TNEE306 pKa = 4.67DD307 pKa = 3.81RR308 pKa = 11.84DD309 pKa = 4.1GDD311 pKa = 3.98GHH313 pKa = 7.3QDD315 pKa = 3.14TKK317 pKa = 11.44DD318 pKa = 3.42NCVEE322 pKa = 4.27IPNSSQLDD330 pKa = 3.68SDD332 pKa = 4.3NDD334 pKa = 4.23GQGDD338 pKa = 4.0DD339 pKa = 5.38CDD341 pKa = 5.28NDD343 pKa = 4.19DD344 pKa = 5.42DD345 pKa = 5.52NDD347 pKa = 5.14GIPDD351 pKa = 4.17YY352 pKa = 10.95LPPGPDD358 pKa = 3.02NCRR361 pKa = 11.84LIANPNQKK369 pKa = 10.35DD370 pKa = 3.34VDD372 pKa = 4.1GNGVGDD378 pKa = 3.67ACEE381 pKa = 3.98EE382 pKa = 4.22DD383 pKa = 3.85FDD385 pKa = 5.35NDD387 pKa = 3.74TVADD391 pKa = 5.0PIDD394 pKa = 3.65VCPEE398 pKa = 3.74SSEE401 pKa = 4.04VTLTDD406 pKa = 3.73FRR408 pKa = 11.84AYY410 pKa = 8.36QTVILDD416 pKa = 3.91PEE418 pKa = 5.2GDD420 pKa = 3.69AQIDD424 pKa = 4.13PNWVVLNQGMEE435 pKa = 4.11IVQTMNSDD443 pKa = 3.19PGLAVGYY450 pKa = 7.31TAFNGVDD457 pKa = 3.99FEE459 pKa = 4.97GTFHH463 pKa = 6.68VNTMTDD469 pKa = 2.95DD470 pKa = 4.36DD471 pKa = 4.35YY472 pKa = 12.02AGFIFAYY479 pKa = 9.53QDD481 pKa = 3.19SASFYY486 pKa = 10.36VVMWKK491 pKa = 7.78QTEE494 pKa = 3.86QTYY497 pKa = 8.13WQATPFRR504 pKa = 11.84AVAEE508 pKa = 4.33SSLQLKK514 pKa = 9.19AVKK517 pKa = 10.44SEE519 pKa = 4.18TGPGEE524 pKa = 4.19YY525 pKa = 10.12LRR527 pKa = 11.84NALWHH532 pKa = 6.17TGHH535 pKa = 6.64TPGHH539 pKa = 6.2VKK541 pKa = 10.49LLWKK545 pKa = 10.39DD546 pKa = 3.35PRR548 pKa = 11.84NVGWKK553 pKa = 10.34DD554 pKa = 2.96KK555 pKa = 10.8TSYY558 pKa = 10.16RR559 pKa = 11.84WRR561 pKa = 11.84LLHH564 pKa = 6.59RR565 pKa = 11.84PQVGYY570 pKa = 10.39IRR572 pKa = 4.61
Molecular weight: 61.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J1W3W9|A0A6J1W3W9_9SAUR serpin B3-like OS=Notechis scutatus OX=8663 GN=LOC113431791 PE=4 SV=1
MM1 pKa = 7.72LKK3 pKa = 10.06NKK5 pKa = 9.86QPIPVNIRR13 pKa = 11.84ATMQQQQLASARR25 pKa = 11.84NRR27 pKa = 11.84RR28 pKa = 11.84LAQQMEE34 pKa = 4.32NRR36 pKa = 11.84PSVQAALKK44 pKa = 10.37LKK46 pKa = 10.39QKK48 pKa = 9.42SLKK51 pKa = 9.72QRR53 pKa = 11.84LGKK56 pKa = 10.77SNIQARR62 pKa = 11.84LGRR65 pKa = 11.84PALARR70 pKa = 11.84GAFGGRR76 pKa = 11.84GLPMGQRR83 pKa = 11.84GLARR87 pKa = 11.84GALARR92 pKa = 11.84GGRR95 pKa = 11.84GARR98 pKa = 11.84GLLRR102 pKa = 11.84GGIPLRR108 pKa = 11.84GHH110 pKa = 5.58SLLRR114 pKa = 11.84GGRR117 pKa = 11.84GLGPRR122 pKa = 11.84MGLRR126 pKa = 11.84RR127 pKa = 11.84GGMRR131 pKa = 11.84GRR133 pKa = 11.84AGPGRR138 pKa = 11.84GGLGRR143 pKa = 11.84GAMGRR148 pKa = 11.84GGLGGRR154 pKa = 11.84GEE156 pKa = 4.31WNGPEE161 pKa = 4.2GQQ163 pKa = 3.58
MM1 pKa = 7.72LKK3 pKa = 10.06NKK5 pKa = 9.86QPIPVNIRR13 pKa = 11.84ATMQQQQLASARR25 pKa = 11.84NRR27 pKa = 11.84RR28 pKa = 11.84LAQQMEE34 pKa = 4.32NRR36 pKa = 11.84PSVQAALKK44 pKa = 10.37LKK46 pKa = 10.39QKK48 pKa = 9.42SLKK51 pKa = 9.72QRR53 pKa = 11.84LGKK56 pKa = 10.77SNIQARR62 pKa = 11.84LGRR65 pKa = 11.84PALARR70 pKa = 11.84GAFGGRR76 pKa = 11.84GLPMGQRR83 pKa = 11.84GLARR87 pKa = 11.84GALARR92 pKa = 11.84GGRR95 pKa = 11.84GARR98 pKa = 11.84GLLRR102 pKa = 11.84GGIPLRR108 pKa = 11.84GHH110 pKa = 5.58SLLRR114 pKa = 11.84GGRR117 pKa = 11.84GLGPRR122 pKa = 11.84MGLRR126 pKa = 11.84RR127 pKa = 11.84GGMRR131 pKa = 11.84GRR133 pKa = 11.84AGPGRR138 pKa = 11.84GGLGRR143 pKa = 11.84GAMGRR148 pKa = 11.84GGLGGRR154 pKa = 11.84GEE156 pKa = 4.31WNGPEE161 pKa = 4.2GQQ163 pKa = 3.58
Molecular weight: 17.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
17436458 |
31 |
33049 |
652.3 |
73.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.345 ± 0.013 | 2.124 ± 0.014 |
4.887 ± 0.01 | 7.526 ± 0.028 |
3.646 ± 0.016 | 5.96 ± 0.017 |
2.512 ± 0.015 | 5.101 ± 0.02 |
6.631 ± 0.034 | 9.321 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.203 ± 0.014 | 4.123 ± 0.016 |
5.833 ± 0.026 | 4.684 ± 0.033 |
5.167 ± 0.012 | 8.417 ± 0.026 |
5.537 ± 0.026 | 6.018 ± 0.038 |
1.154 ± 0.005 | 2.807 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
