
Bos taurus papillomavirus 4 (Bovine papillomavirus 4)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Xipapillomavirus; Xipapillomavirus 1
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
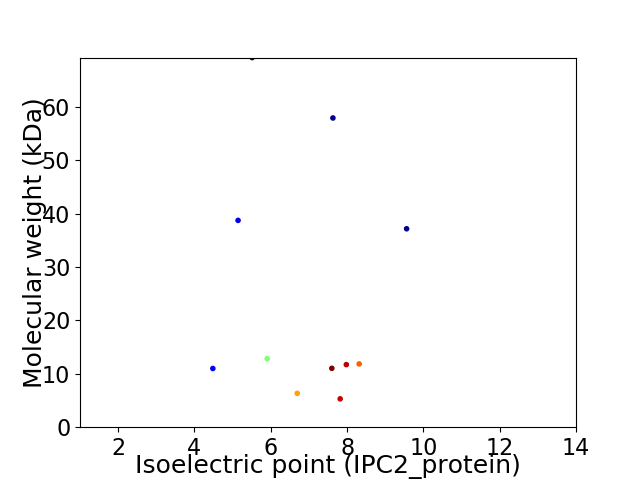
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P08352|VE8_BPV4 Uncharacterized protein E8 OS=Bos taurus papillomavirus 4 OX=10562 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 9.93GQNVTLQDD10 pKa = 3.25IAIEE14 pKa = 4.24LEE16 pKa = 4.3DD17 pKa = 4.44TISPINLHH25 pKa = 6.07CEE27 pKa = 4.02EE28 pKa = 4.83EE29 pKa = 4.44IEE31 pKa = 4.28TEE33 pKa = 4.69EE34 pKa = 4.45VDD36 pKa = 3.83TPNPFAITATCYY48 pKa = 10.59ACEE51 pKa = 3.88QVLRR55 pKa = 11.84LAVVTSTEE63 pKa = 4.71GIHH66 pKa = 6.57QLQQLLFDD74 pKa = 5.02NLFLLCAACSKK85 pKa = 10.58QVFCNRR91 pKa = 11.84RR92 pKa = 11.84PEE94 pKa = 4.16RR95 pKa = 11.84NGPP98 pKa = 3.49
MM1 pKa = 7.63KK2 pKa = 9.93GQNVTLQDD10 pKa = 3.25IAIEE14 pKa = 4.24LEE16 pKa = 4.3DD17 pKa = 4.44TISPINLHH25 pKa = 6.07CEE27 pKa = 4.02EE28 pKa = 4.83EE29 pKa = 4.44IEE31 pKa = 4.28TEE33 pKa = 4.69EE34 pKa = 4.45VDD36 pKa = 3.83TPNPFAITATCYY48 pKa = 10.59ACEE51 pKa = 3.88QVLRR55 pKa = 11.84LAVVTSTEE63 pKa = 4.71GIHH66 pKa = 6.57QLQQLLFDD74 pKa = 5.02NLFLLCAACSKK85 pKa = 10.58QVFCNRR91 pKa = 11.84RR92 pKa = 11.84PEE94 pKa = 4.16RR95 pKa = 11.84NGPP98 pKa = 3.49
Molecular weight: 10.99 kDa
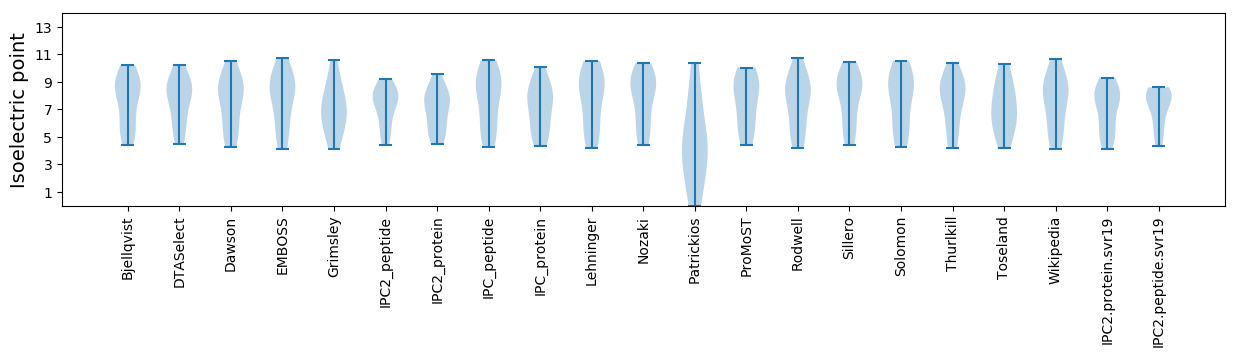
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P08346|VE3_BPV4 Probable protein E3 OS=Bos taurus papillomavirus 4 OX=10562 GN=E3 PE=4 SV=1
MM1 pKa = 6.96YY2 pKa = 10.19LCLEE6 pKa = 4.29SLQKK10 pKa = 10.58SEE12 pKa = 4.92FANQRR17 pKa = 11.84WSLVDD22 pKa = 3.14TSIEE26 pKa = 4.16TFKK29 pKa = 11.23APPEE33 pKa = 3.98NTLKK37 pKa = 10.77KK38 pKa = 9.91RR39 pKa = 11.84GQHH42 pKa = 3.92VTVIYY47 pKa = 9.9DD48 pKa = 3.56QNAMNSMVYY57 pKa = 8.48TLWKK61 pKa = 8.63EE62 pKa = 3.91VYY64 pKa = 10.01YY65 pKa = 10.94VDD67 pKa = 5.17EE68 pKa = 4.57TEE70 pKa = 4.27TWHH73 pKa = 6.09KK74 pKa = 9.6TSSDD78 pKa = 3.38LDD80 pKa = 3.71YY81 pKa = 11.71DD82 pKa = 4.2GIFYY86 pKa = 10.2IDD88 pKa = 3.42NQGNKK93 pKa = 8.64IYY95 pKa = 10.66YY96 pKa = 10.65VNFQDD101 pKa = 5.78DD102 pKa = 3.48AALYY106 pKa = 9.36SNSGMGQVHH115 pKa = 6.57FEE117 pKa = 4.41SKK119 pKa = 10.53VLSPSVTSSLRR130 pKa = 11.84VGSTGGQRR138 pKa = 11.84GTQTGDD144 pKa = 2.82HH145 pKa = 6.16ARR147 pKa = 11.84GRR149 pKa = 11.84SRR151 pKa = 11.84PSPRR155 pKa = 11.84SSRR158 pKa = 11.84DD159 pKa = 2.95ARR161 pKa = 11.84GRR163 pKa = 11.84QQRR166 pKa = 11.84AQSSSRR172 pKa = 11.84SRR174 pKa = 11.84SRR176 pKa = 11.84SRR178 pKa = 11.84SRR180 pKa = 11.84SPTKK184 pKa = 10.42GPHH187 pKa = 5.18SSGRR191 pKa = 11.84DD192 pKa = 3.18TRR194 pKa = 11.84LPSPGRR200 pKa = 11.84PPGGRR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84GTPEE211 pKa = 3.79RR212 pKa = 11.84EE213 pKa = 4.05RR214 pKa = 11.84CPGTPTPPTPDD225 pKa = 3.02QVGGRR230 pKa = 11.84SSTPKK235 pKa = 9.6RR236 pKa = 11.84QASSRR241 pKa = 11.84LAQLIDD247 pKa = 3.43AAYY250 pKa = 9.68DD251 pKa = 3.66PPVLLLQGAANTLKK265 pKa = 10.47CFRR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84ATQAHH275 pKa = 5.24PHH277 pKa = 6.58KK278 pKa = 9.05FLCMSTSWTWVSKK291 pKa = 9.41TSPLKK296 pKa = 10.3SGHH299 pKa = 6.3RR300 pKa = 11.84MLIAFSNSEE309 pKa = 3.65QRR311 pKa = 11.84NCFLASVRR319 pKa = 11.84LPKK322 pKa = 10.5GVSAVKK328 pKa = 10.33GALDD332 pKa = 3.52GLL334 pKa = 4.31
MM1 pKa = 6.96YY2 pKa = 10.19LCLEE6 pKa = 4.29SLQKK10 pKa = 10.58SEE12 pKa = 4.92FANQRR17 pKa = 11.84WSLVDD22 pKa = 3.14TSIEE26 pKa = 4.16TFKK29 pKa = 11.23APPEE33 pKa = 3.98NTLKK37 pKa = 10.77KK38 pKa = 9.91RR39 pKa = 11.84GQHH42 pKa = 3.92VTVIYY47 pKa = 9.9DD48 pKa = 3.56QNAMNSMVYY57 pKa = 8.48TLWKK61 pKa = 8.63EE62 pKa = 3.91VYY64 pKa = 10.01YY65 pKa = 10.94VDD67 pKa = 5.17EE68 pKa = 4.57TEE70 pKa = 4.27TWHH73 pKa = 6.09KK74 pKa = 9.6TSSDD78 pKa = 3.38LDD80 pKa = 3.71YY81 pKa = 11.71DD82 pKa = 4.2GIFYY86 pKa = 10.2IDD88 pKa = 3.42NQGNKK93 pKa = 8.64IYY95 pKa = 10.66YY96 pKa = 10.65VNFQDD101 pKa = 5.78DD102 pKa = 3.48AALYY106 pKa = 9.36SNSGMGQVHH115 pKa = 6.57FEE117 pKa = 4.41SKK119 pKa = 10.53VLSPSVTSSLRR130 pKa = 11.84VGSTGGQRR138 pKa = 11.84GTQTGDD144 pKa = 2.82HH145 pKa = 6.16ARR147 pKa = 11.84GRR149 pKa = 11.84SRR151 pKa = 11.84PSPRR155 pKa = 11.84SSRR158 pKa = 11.84DD159 pKa = 2.95ARR161 pKa = 11.84GRR163 pKa = 11.84QQRR166 pKa = 11.84AQSSSRR172 pKa = 11.84SRR174 pKa = 11.84SRR176 pKa = 11.84SRR178 pKa = 11.84SRR180 pKa = 11.84SPTKK184 pKa = 10.42GPHH187 pKa = 5.18SSGRR191 pKa = 11.84DD192 pKa = 3.18TRR194 pKa = 11.84LPSPGRR200 pKa = 11.84PPGGRR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84GTPEE211 pKa = 3.79RR212 pKa = 11.84EE213 pKa = 4.05RR214 pKa = 11.84CPGTPTPPTPDD225 pKa = 3.02QVGGRR230 pKa = 11.84SSTPKK235 pKa = 9.6RR236 pKa = 11.84QASSRR241 pKa = 11.84LAQLIDD247 pKa = 3.43AAYY250 pKa = 9.68DD251 pKa = 3.66PPVLLLQGAANTLKK265 pKa = 10.47CFRR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84ATQAHH275 pKa = 5.24PHH277 pKa = 6.58KK278 pKa = 9.05FLCMSTSWTWVSKK291 pKa = 9.41TSPLKK296 pKa = 10.3SGHH299 pKa = 6.3RR300 pKa = 11.84MLIAFSNSEE309 pKa = 3.65QRR311 pKa = 11.84NCFLASVRR319 pKa = 11.84LPKK322 pKa = 10.5GVSAVKK328 pKa = 10.33GALDD332 pKa = 3.52GLL334 pKa = 4.31
Molecular weight: 37.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2416 |
48 |
608 |
219.6 |
24.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.96 ± 0.502 | 2.649 ± 0.545 |
5.877 ± 0.468 | 5.422 ± 0.619 |
4.636 ± 0.574 | 6.25 ± 0.701 |
2.152 ± 0.178 | 4.346 ± 0.589 |
5.132 ± 0.586 | 8.982 ± 1.186 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.945 ± 0.207 | 4.222 ± 0.469 |
6.333 ± 0.737 | 4.967 ± 0.34 |
6.581 ± 0.933 | 7.326 ± 0.978 |
6.705 ± 0.419 | 5.712 ± 0.415 |
1.697 ± 0.277 | 3.104 ± 0.466 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
