
Domibacillus epiphyticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Domibacillus
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
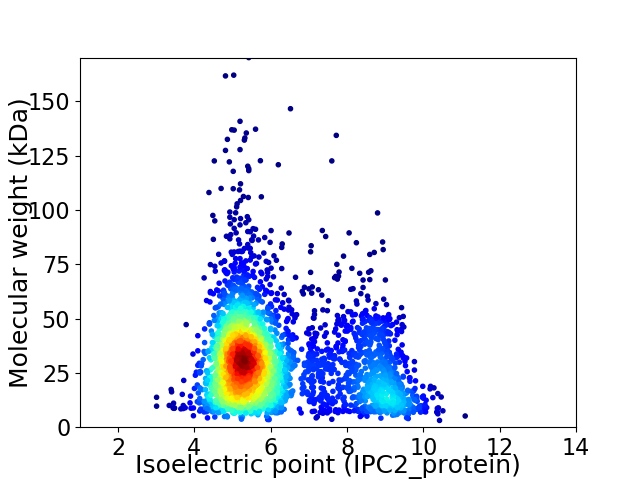
Virtual 2D-PAGE plot for 3171 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V2A6U5|A0A1V2A6U5_9BACI HD domain-containing protein OS=Domibacillus epiphyticus OX=1714355 GN=BTO28_10895 PE=4 SV=1
MM1 pKa = 7.16YY2 pKa = 10.42CSTCGTNLGRR12 pKa = 11.84SSRR15 pKa = 11.84YY16 pKa = 9.83CPGCGSKK23 pKa = 10.1RR24 pKa = 11.84GRR26 pKa = 11.84RR27 pKa = 11.84FSYY30 pKa = 10.46VKK32 pKa = 10.53LIAFLCIASVLAFAGTIYY50 pKa = 11.02YY51 pKa = 10.57LLSNDD56 pKa = 4.39EE57 pKa = 4.27KK58 pKa = 10.51TQTASQTAIKK68 pKa = 9.55TPTEE72 pKa = 3.77QSADD76 pKa = 3.39TRR78 pKa = 11.84KK79 pKa = 7.58TVAPPEE85 pKa = 4.11EE86 pKa = 4.84LSAEE90 pKa = 4.23DD91 pKa = 3.66QQSLTEE97 pKa = 4.72IIAAAQQNVYY107 pKa = 10.14TIFTDD112 pKa = 3.34SGQGSGFLINDD123 pKa = 3.87RR124 pKa = 11.84GDD126 pKa = 3.43VATNAHH132 pKa = 5.11VVEE135 pKa = 4.92GSVSVIVKK143 pKa = 10.44DD144 pKa = 3.34IDD146 pKa = 3.91GVEE149 pKa = 4.19HH150 pKa = 7.01EE151 pKa = 4.86GTVIGYY157 pKa = 8.66SNKK160 pKa = 9.36TDD162 pKa = 3.11VALIRR167 pKa = 11.84VPDD170 pKa = 3.97FADD173 pKa = 3.54YY174 pKa = 11.35SPLNLEE180 pKa = 4.4TNKK183 pKa = 10.1KK184 pKa = 9.29AQVGDD189 pKa = 3.67EE190 pKa = 4.4VIALGSPLALEE201 pKa = 4.12NTATFGYY208 pKa = 8.06ITGVDD213 pKa = 2.9RR214 pKa = 11.84SFIIEE219 pKa = 3.86PHH221 pKa = 6.47SYY223 pKa = 11.41DD224 pKa = 4.55NIYY227 pKa = 8.18QTSAAIAPGSSGGPLVSKK245 pKa = 9.57NTGRR249 pKa = 11.84VIAINSAKK257 pKa = 9.69LTGEE261 pKa = 3.94DD262 pKa = 4.08AIGFSIPYY270 pKa = 9.77KK271 pKa = 10.8DD272 pKa = 3.97VDD274 pKa = 4.45DD275 pKa = 4.44MLKK278 pKa = 9.74KK279 pKa = 10.04WSRR282 pKa = 11.84SPLSEE287 pKa = 4.38EE288 pKa = 3.93EE289 pKa = 4.35VNDD292 pKa = 4.29LFYY295 pKa = 11.09TEE297 pKa = 5.33DD298 pKa = 3.37GSLFFEE304 pKa = 4.96EE305 pKa = 5.08YY306 pKa = 9.54WNEE309 pKa = 3.64DD310 pKa = 3.15AYY312 pKa = 11.33FDD314 pKa = 4.05GGEE317 pKa = 4.11YY318 pKa = 10.36SDD320 pKa = 4.22DD321 pKa = 3.61ASYY324 pKa = 11.73DD325 pKa = 3.71FYY327 pKa = 11.25DD328 pKa = 5.19IPADD332 pKa = 2.78WWYY335 pKa = 10.7EE336 pKa = 3.49EE337 pKa = 4.05DD338 pKa = 3.96TYY340 pKa = 11.43EE341 pKa = 4.96DD342 pKa = 4.32EE343 pKa = 4.96YY344 pKa = 11.11IEE346 pKa = 4.17EE347 pKa = 4.32VPEE350 pKa = 3.92EE351 pKa = 4.69EE352 pKa = 4.84IPSTEE357 pKa = 3.75PDD359 pKa = 3.55DD360 pKa = 4.69YY361 pKa = 11.63EE362 pKa = 4.3EE363 pKa = 4.12EE364 pKa = 4.46TIEE367 pKa = 4.04EE368 pKa = 4.29EE369 pKa = 4.69PIEE372 pKa = 4.25EE373 pKa = 4.21LPLIEE378 pKa = 5.04EE379 pKa = 4.48EE380 pKa = 4.58PMPDD384 pKa = 3.63TAPDD388 pKa = 4.23PYY390 pKa = 11.26LEE392 pKa = 5.53DD393 pKa = 3.51INGDD397 pKa = 3.61GLIDD401 pKa = 3.91VNDD404 pKa = 3.53VTEE407 pKa = 5.32DD408 pKa = 3.18INMDD412 pKa = 3.69GVIDD416 pKa = 4.31EE417 pKa = 5.28LDD419 pKa = 3.74LDD421 pKa = 4.65EE422 pKa = 6.2LLMMQQ427 pKa = 4.84
MM1 pKa = 7.16YY2 pKa = 10.42CSTCGTNLGRR12 pKa = 11.84SSRR15 pKa = 11.84YY16 pKa = 9.83CPGCGSKK23 pKa = 10.1RR24 pKa = 11.84GRR26 pKa = 11.84RR27 pKa = 11.84FSYY30 pKa = 10.46VKK32 pKa = 10.53LIAFLCIASVLAFAGTIYY50 pKa = 11.02YY51 pKa = 10.57LLSNDD56 pKa = 4.39EE57 pKa = 4.27KK58 pKa = 10.51TQTASQTAIKK68 pKa = 9.55TPTEE72 pKa = 3.77QSADD76 pKa = 3.39TRR78 pKa = 11.84KK79 pKa = 7.58TVAPPEE85 pKa = 4.11EE86 pKa = 4.84LSAEE90 pKa = 4.23DD91 pKa = 3.66QQSLTEE97 pKa = 4.72IIAAAQQNVYY107 pKa = 10.14TIFTDD112 pKa = 3.34SGQGSGFLINDD123 pKa = 3.87RR124 pKa = 11.84GDD126 pKa = 3.43VATNAHH132 pKa = 5.11VVEE135 pKa = 4.92GSVSVIVKK143 pKa = 10.44DD144 pKa = 3.34IDD146 pKa = 3.91GVEE149 pKa = 4.19HH150 pKa = 7.01EE151 pKa = 4.86GTVIGYY157 pKa = 8.66SNKK160 pKa = 9.36TDD162 pKa = 3.11VALIRR167 pKa = 11.84VPDD170 pKa = 3.97FADD173 pKa = 3.54YY174 pKa = 11.35SPLNLEE180 pKa = 4.4TNKK183 pKa = 10.1KK184 pKa = 9.29AQVGDD189 pKa = 3.67EE190 pKa = 4.4VIALGSPLALEE201 pKa = 4.12NTATFGYY208 pKa = 8.06ITGVDD213 pKa = 2.9RR214 pKa = 11.84SFIIEE219 pKa = 3.86PHH221 pKa = 6.47SYY223 pKa = 11.41DD224 pKa = 4.55NIYY227 pKa = 8.18QTSAAIAPGSSGGPLVSKK245 pKa = 9.57NTGRR249 pKa = 11.84VIAINSAKK257 pKa = 9.69LTGEE261 pKa = 3.94DD262 pKa = 4.08AIGFSIPYY270 pKa = 9.77KK271 pKa = 10.8DD272 pKa = 3.97VDD274 pKa = 4.45DD275 pKa = 4.44MLKK278 pKa = 9.74KK279 pKa = 10.04WSRR282 pKa = 11.84SPLSEE287 pKa = 4.38EE288 pKa = 3.93EE289 pKa = 4.35VNDD292 pKa = 4.29LFYY295 pKa = 11.09TEE297 pKa = 5.33DD298 pKa = 3.37GSLFFEE304 pKa = 4.96EE305 pKa = 5.08YY306 pKa = 9.54WNEE309 pKa = 3.64DD310 pKa = 3.15AYY312 pKa = 11.33FDD314 pKa = 4.05GGEE317 pKa = 4.11YY318 pKa = 10.36SDD320 pKa = 4.22DD321 pKa = 3.61ASYY324 pKa = 11.73DD325 pKa = 3.71FYY327 pKa = 11.25DD328 pKa = 5.19IPADD332 pKa = 2.78WWYY335 pKa = 10.7EE336 pKa = 3.49EE337 pKa = 4.05DD338 pKa = 3.96TYY340 pKa = 11.43EE341 pKa = 4.96DD342 pKa = 4.32EE343 pKa = 4.96YY344 pKa = 11.11IEE346 pKa = 4.17EE347 pKa = 4.32VPEE350 pKa = 3.92EE351 pKa = 4.69EE352 pKa = 4.84IPSTEE357 pKa = 3.75PDD359 pKa = 3.55DD360 pKa = 4.69YY361 pKa = 11.63EE362 pKa = 4.3EE363 pKa = 4.12EE364 pKa = 4.46TIEE367 pKa = 4.04EE368 pKa = 4.29EE369 pKa = 4.69PIEE372 pKa = 4.25EE373 pKa = 4.21LPLIEE378 pKa = 5.04EE379 pKa = 4.48EE380 pKa = 4.58PMPDD384 pKa = 3.63TAPDD388 pKa = 4.23PYY390 pKa = 11.26LEE392 pKa = 5.53DD393 pKa = 3.51INGDD397 pKa = 3.61GLIDD401 pKa = 3.91VNDD404 pKa = 3.53VTEE407 pKa = 5.32DD408 pKa = 3.18INMDD412 pKa = 3.69GVIDD416 pKa = 4.31EE417 pKa = 5.28LDD419 pKa = 3.74LDD421 pKa = 4.65EE422 pKa = 6.2LLMMQQ427 pKa = 4.84
Molecular weight: 47.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V2A6M7|A0A1V2A6M7_9BACI MFS transporter OS=Domibacillus epiphyticus OX=1714355 GN=BTO28_12375 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.21RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84SKK14 pKa = 9.83VHH16 pKa = 5.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSKK25 pKa = 9.98NGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.74VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.21RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84SKK14 pKa = 9.83VHH16 pKa = 5.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSKK25 pKa = 9.98NGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.74VLSAA44 pKa = 4.05
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
908978 |
27 |
1529 |
286.7 |
32.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.866 ± 0.047 | 0.719 ± 0.014 |
5.169 ± 0.031 | 7.622 ± 0.052 |
4.503 ± 0.036 | 7.144 ± 0.039 |
2.117 ± 0.021 | 7.518 ± 0.044 |
6.582 ± 0.042 | 9.418 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.935 ± 0.021 | 4.05 ± 0.031 |
3.769 ± 0.027 | 3.451 ± 0.027 |
4.319 ± 0.032 | 6.024 ± 0.031 |
5.402 ± 0.03 | 7.143 ± 0.032 |
0.976 ± 0.016 | 3.273 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
