
Natrarchaeobaculum sulfurireducens
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natrarchaeobaculum
Average proteome isoelectric point is 4.86
Get precalculated fractions of proteins
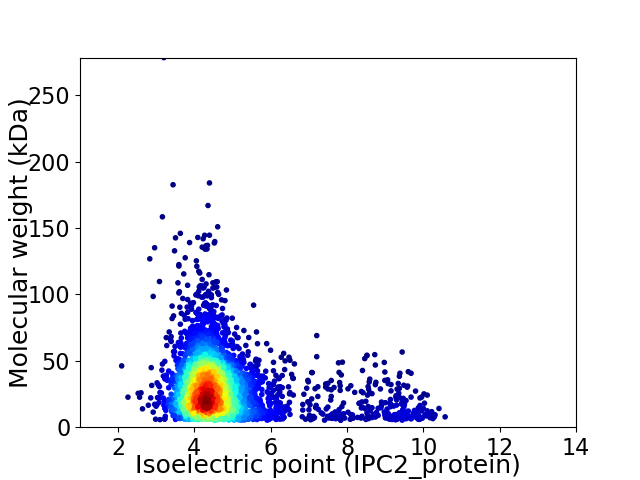
Virtual 2D-PAGE plot for 3734 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346PQM8|A0A346PQM8_9EURY Uncharacterized protein OS=Natrarchaeobaculum sulfurireducens OX=2044521 GN=AArcMg_1816 PE=4 SV=1
MM1 pKa = 7.49SDD3 pKa = 2.89EE4 pKa = 3.86RR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 10.53ASRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.74LLAATGSLTALGLAGCLGDD31 pKa = 4.26EE32 pKa = 4.89EE33 pKa = 5.75GDD35 pKa = 3.81EE36 pKa = 4.82PEE38 pKa = 4.78PADD41 pKa = 5.94DD42 pKa = 6.13GDD44 pKa = 5.37DD45 pKa = 3.98DD46 pKa = 6.42HH47 pKa = 9.36DD48 pKa = 6.87DD49 pKa = 4.85DD50 pKa = 6.27DD51 pKa = 5.85HH52 pKa = 9.08DD53 pKa = 5.79HH54 pKa = 7.79DD55 pKa = 6.51DD56 pKa = 5.3DD57 pKa = 5.59HH58 pKa = 8.97DD59 pKa = 5.46HH60 pKa = 7.78DD61 pKa = 6.31DD62 pKa = 4.16GVADD66 pKa = 4.58EE67 pKa = 5.24YY68 pKa = 11.4FDD70 pKa = 3.9EE71 pKa = 6.02LGVADD76 pKa = 4.72EE77 pKa = 5.14GFVILDD83 pKa = 3.68RR84 pKa = 11.84AHH86 pKa = 7.2DD87 pKa = 3.67PHH89 pKa = 8.28EE90 pKa = 4.33EE91 pKa = 3.7VAYY94 pKa = 9.99VHH96 pKa = 6.84GDD98 pKa = 2.82HH99 pKa = 6.46WHH101 pKa = 6.98GDD103 pKa = 3.65LPSIPVDD110 pKa = 3.7DD111 pKa = 4.34NVSIGAEE118 pKa = 3.89IEE120 pKa = 4.76LEE122 pKa = 4.58DD123 pKa = 5.02GDD125 pKa = 4.34EE126 pKa = 4.59LEE128 pKa = 5.96LGDD131 pKa = 4.14EE132 pKa = 4.48YY133 pKa = 11.14EE134 pKa = 4.39LRR136 pKa = 11.84VAVAPDD142 pKa = 3.46EE143 pKa = 4.52PEE145 pKa = 4.03GVVGIDD151 pKa = 3.04EE152 pKa = 4.89DD153 pKa = 5.36DD154 pKa = 3.46YY155 pKa = 11.78HH156 pKa = 7.73GDD158 pKa = 3.29HH159 pKa = 5.82VHH161 pKa = 6.09IHH163 pKa = 6.36GEE165 pKa = 4.1QEE167 pKa = 4.12GVTEE171 pKa = 4.25VVFMIWHH178 pKa = 7.56DD179 pKa = 3.72DD180 pKa = 3.48HH181 pKa = 9.02ADD183 pKa = 3.67YY184 pKa = 10.87QSPPITAQVVEE195 pKa = 4.56EE196 pKa = 5.19DD197 pKa = 4.31DD198 pKa = 5.52DD199 pKa = 5.14HH200 pKa = 8.92DD201 pKa = 4.84HH202 pKa = 7.4DD203 pKa = 6.05HH204 pKa = 7.4DD205 pKa = 4.52DD206 pKa = 3.92EE207 pKa = 4.69YY208 pKa = 11.94VSEE211 pKa = 4.31FQLLDD216 pKa = 3.58RR217 pKa = 11.84AHH219 pKa = 7.27DD220 pKa = 3.57PHH222 pKa = 8.4EE223 pKa = 4.86EE224 pKa = 3.41ISYY227 pKa = 8.76VHH229 pKa = 6.82GDD231 pKa = 3.02HH232 pKa = 6.56WHH234 pKa = 6.74GEE236 pKa = 4.14DD237 pKa = 4.97DD238 pKa = 4.5FPTIPVGDD246 pKa = 3.86NVSIGAEE253 pKa = 4.08LEE255 pKa = 4.24DD256 pKa = 4.31ADD258 pKa = 4.49GDD260 pKa = 4.4EE261 pKa = 4.53IVLGPEE267 pKa = 3.85YY268 pKa = 10.65EE269 pKa = 4.43LGVEE273 pKa = 4.14LADD276 pKa = 3.96GAEE279 pKa = 4.08EE280 pKa = 4.42GVVEE284 pKa = 4.13IDD286 pKa = 3.85EE287 pKa = 5.15DD288 pKa = 3.85EE289 pKa = 5.44DD290 pKa = 3.89YY291 pKa = 11.42HH292 pKa = 8.16GDD294 pKa = 3.4HH295 pKa = 5.84VHH297 pKa = 6.05IHH299 pKa = 6.3GEE301 pKa = 4.25SAGEE305 pKa = 4.15TEE307 pKa = 4.85VVFTLIHH314 pKa = 7.19DD315 pKa = 3.82DD316 pKa = 3.61HH317 pKa = 8.73VDD319 pKa = 3.59YY320 pKa = 7.47EE321 pKa = 4.89TPALTVTVGDD331 pKa = 4.27DD332 pKa = 3.05
MM1 pKa = 7.49SDD3 pKa = 2.89EE4 pKa = 3.86RR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 10.53ASRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.74LLAATGSLTALGLAGCLGDD31 pKa = 4.26EE32 pKa = 4.89EE33 pKa = 5.75GDD35 pKa = 3.81EE36 pKa = 4.82PEE38 pKa = 4.78PADD41 pKa = 5.94DD42 pKa = 6.13GDD44 pKa = 5.37DD45 pKa = 3.98DD46 pKa = 6.42HH47 pKa = 9.36DD48 pKa = 6.87DD49 pKa = 4.85DD50 pKa = 6.27DD51 pKa = 5.85HH52 pKa = 9.08DD53 pKa = 5.79HH54 pKa = 7.79DD55 pKa = 6.51DD56 pKa = 5.3DD57 pKa = 5.59HH58 pKa = 8.97DD59 pKa = 5.46HH60 pKa = 7.78DD61 pKa = 6.31DD62 pKa = 4.16GVADD66 pKa = 4.58EE67 pKa = 5.24YY68 pKa = 11.4FDD70 pKa = 3.9EE71 pKa = 6.02LGVADD76 pKa = 4.72EE77 pKa = 5.14GFVILDD83 pKa = 3.68RR84 pKa = 11.84AHH86 pKa = 7.2DD87 pKa = 3.67PHH89 pKa = 8.28EE90 pKa = 4.33EE91 pKa = 3.7VAYY94 pKa = 9.99VHH96 pKa = 6.84GDD98 pKa = 2.82HH99 pKa = 6.46WHH101 pKa = 6.98GDD103 pKa = 3.65LPSIPVDD110 pKa = 3.7DD111 pKa = 4.34NVSIGAEE118 pKa = 3.89IEE120 pKa = 4.76LEE122 pKa = 4.58DD123 pKa = 5.02GDD125 pKa = 4.34EE126 pKa = 4.59LEE128 pKa = 5.96LGDD131 pKa = 4.14EE132 pKa = 4.48YY133 pKa = 11.14EE134 pKa = 4.39LRR136 pKa = 11.84VAVAPDD142 pKa = 3.46EE143 pKa = 4.52PEE145 pKa = 4.03GVVGIDD151 pKa = 3.04EE152 pKa = 4.89DD153 pKa = 5.36DD154 pKa = 3.46YY155 pKa = 11.78HH156 pKa = 7.73GDD158 pKa = 3.29HH159 pKa = 5.82VHH161 pKa = 6.09IHH163 pKa = 6.36GEE165 pKa = 4.1QEE167 pKa = 4.12GVTEE171 pKa = 4.25VVFMIWHH178 pKa = 7.56DD179 pKa = 3.72DD180 pKa = 3.48HH181 pKa = 9.02ADD183 pKa = 3.67YY184 pKa = 10.87QSPPITAQVVEE195 pKa = 4.56EE196 pKa = 5.19DD197 pKa = 4.31DD198 pKa = 5.52DD199 pKa = 5.14HH200 pKa = 8.92DD201 pKa = 4.84HH202 pKa = 7.4DD203 pKa = 6.05HH204 pKa = 7.4DD205 pKa = 4.52DD206 pKa = 3.92EE207 pKa = 4.69YY208 pKa = 11.94VSEE211 pKa = 4.31FQLLDD216 pKa = 3.58RR217 pKa = 11.84AHH219 pKa = 7.27DD220 pKa = 3.57PHH222 pKa = 8.4EE223 pKa = 4.86EE224 pKa = 3.41ISYY227 pKa = 8.76VHH229 pKa = 6.82GDD231 pKa = 3.02HH232 pKa = 6.56WHH234 pKa = 6.74GEE236 pKa = 4.14DD237 pKa = 4.97DD238 pKa = 4.5FPTIPVGDD246 pKa = 3.86NVSIGAEE253 pKa = 4.08LEE255 pKa = 4.24DD256 pKa = 4.31ADD258 pKa = 4.49GDD260 pKa = 4.4EE261 pKa = 4.53IVLGPEE267 pKa = 3.85YY268 pKa = 10.65EE269 pKa = 4.43LGVEE273 pKa = 4.14LADD276 pKa = 3.96GAEE279 pKa = 4.08EE280 pKa = 4.42GVVEE284 pKa = 4.13IDD286 pKa = 3.85EE287 pKa = 5.15DD288 pKa = 3.85EE289 pKa = 5.44DD290 pKa = 3.89YY291 pKa = 11.42HH292 pKa = 8.16GDD294 pKa = 3.4HH295 pKa = 5.84VHH297 pKa = 6.05IHH299 pKa = 6.3GEE301 pKa = 4.25SAGEE305 pKa = 4.15TEE307 pKa = 4.85VVFTLIHH314 pKa = 7.19DD315 pKa = 3.82DD316 pKa = 3.61HH317 pKa = 8.73VDD319 pKa = 3.59YY320 pKa = 7.47EE321 pKa = 4.89TPALTVTVGDD331 pKa = 4.27DD332 pKa = 3.05
Molecular weight: 36.77 kDa
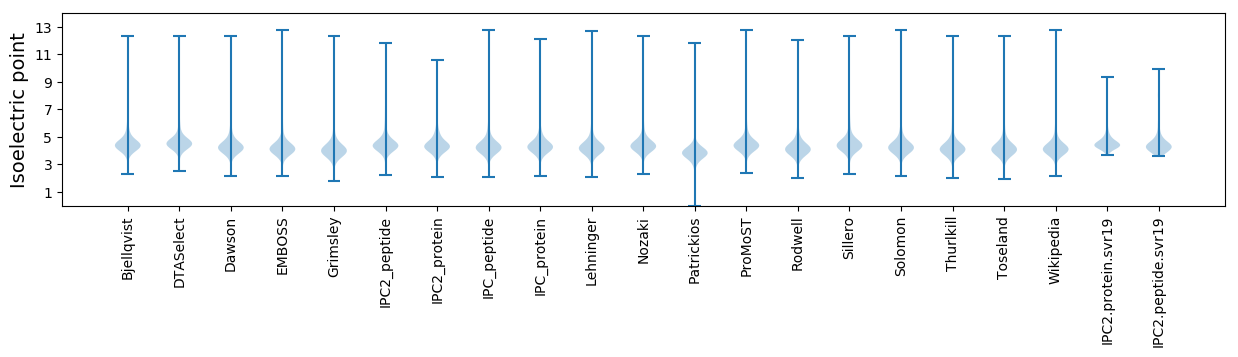
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346PSJ3|A0A346PSJ3_9EURY Phage protein OS=Natrarchaeobaculum sulfurireducens OX=2044521 GN=AArc1_1230 PE=3 SV=1
MM1 pKa = 8.01PYY3 pKa = 10.03RR4 pKa = 11.84VALEE8 pKa = 3.88KK9 pKa = 10.89LVFDD13 pKa = 4.21RR14 pKa = 11.84FGRR17 pKa = 11.84PIVRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84APVPDD28 pKa = 3.9TNSLIDD34 pKa = 3.38ASRR37 pKa = 11.84FLVRR41 pKa = 11.84VGSTDD46 pKa = 3.41EE47 pKa = 4.09PSEE50 pKa = 4.29PFDD53 pKa = 4.15SRR55 pKa = 11.84RR56 pKa = 11.84PTT58 pKa = 3.17
MM1 pKa = 8.01PYY3 pKa = 10.03RR4 pKa = 11.84VALEE8 pKa = 3.88KK9 pKa = 10.89LVFDD13 pKa = 4.21RR14 pKa = 11.84FGRR17 pKa = 11.84PIVRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84APVPDD28 pKa = 3.9TNSLIDD34 pKa = 3.38ASRR37 pKa = 11.84FLVRR41 pKa = 11.84VGSTDD46 pKa = 3.41EE47 pKa = 4.09PSEE50 pKa = 4.29PFDD53 pKa = 4.15SRR55 pKa = 11.84RR56 pKa = 11.84PTT58 pKa = 3.17
Molecular weight: 6.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1078029 |
44 |
2660 |
288.7 |
31.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.455 ± 0.053 | 0.766 ± 0.014 |
8.75 ± 0.057 | 9.244 ± 0.05 |
3.228 ± 0.027 | 8.137 ± 0.038 |
2.033 ± 0.02 | 4.385 ± 0.031 |
1.779 ± 0.026 | 8.93 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.729 ± 0.017 | 2.243 ± 0.021 |
4.557 ± 0.028 | 2.456 ± 0.02 |
6.42 ± 0.048 | 5.596 ± 0.033 |
6.491 ± 0.031 | 9.026 ± 0.042 |
1.112 ± 0.016 | 2.664 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
