
Siphoviridae sp. ct7UA22
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
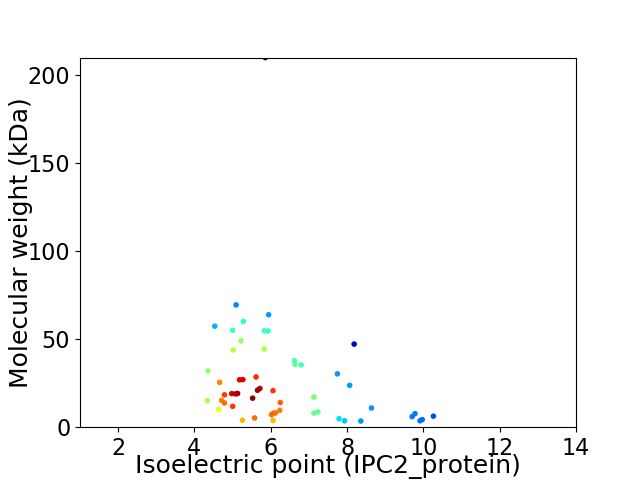
Virtual 2D-PAGE plot for 55 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2WBG7|A0A5Q2WBG7_9CAUD Uncharacterized protein OS=Siphoviridae sp. ct7UA22 OX=2656718 PE=4 SV=1
MM1 pKa = 7.25EE2 pKa = 5.54LEE4 pKa = 4.31IGSLHH9 pKa = 6.75SGGLVVGVNPLEE21 pKa = 4.16LAPNVFNDD29 pKa = 3.32VANVEE34 pKa = 4.55FTRR37 pKa = 11.84EE38 pKa = 3.82GVRR41 pKa = 11.84PLRR44 pKa = 11.84QPVTRR49 pKa = 11.84NLDD52 pKa = 3.5VPGSRR57 pKa = 11.84LVYY60 pKa = 9.96HH61 pKa = 7.07AVDD64 pKa = 3.3TMVNGGIRR72 pKa = 11.84VVLCFCEE79 pKa = 4.2DD80 pKa = 2.59KK81 pKa = 11.23VYY83 pKa = 11.09ALRR86 pKa = 11.84GSSYY90 pKa = 11.33LDD92 pKa = 3.22VTPAGMLSSKK102 pKa = 9.09EE103 pKa = 3.8WSVTEE108 pKa = 3.76FNGYY112 pKa = 8.47VVACNGKK119 pKa = 8.84NFPYY123 pKa = 10.62YY124 pKa = 10.71YY125 pKa = 10.54DD126 pKa = 3.71FADD129 pKa = 4.13ADD131 pKa = 4.22SVLKK135 pKa = 10.34ILPEE139 pKa = 3.86WPEE142 pKa = 3.52EE143 pKa = 3.8LVPQKK148 pKa = 10.39LSSMSGFLVFMGLVSDD164 pKa = 4.13GTFANQLIVWSDD176 pKa = 3.18AAEE179 pKa = 4.45LGSLPDD185 pKa = 3.53NYY187 pKa = 10.99NWADD191 pKa = 3.42PTSRR195 pKa = 11.84AGFYY199 pKa = 10.48SLPDD203 pKa = 3.49FEE205 pKa = 5.24EE206 pKa = 4.45FVAAVLLNKK215 pKa = 10.29SLIIYY220 pKa = 7.46RR221 pKa = 11.84TNSIYY226 pKa = 10.64EE227 pKa = 3.66MRR229 pKa = 11.84FIGGTLVFSIEE240 pKa = 3.69QRR242 pKa = 11.84FEE244 pKa = 3.87GKK246 pKa = 7.95TLISSKK252 pKa = 10.75AVAAKK257 pKa = 10.38GRR259 pKa = 11.84LHH261 pKa = 5.94YY262 pKa = 10.71CIGEE266 pKa = 4.08NEE268 pKa = 4.6FYY270 pKa = 11.41VHH272 pKa = 7.34DD273 pKa = 4.81GSSEE277 pKa = 3.96QVFDD281 pKa = 3.66VRR283 pKa = 11.84PVSDD287 pKa = 4.08YY288 pKa = 11.53YY289 pKa = 11.57FNTVNPDD296 pKa = 3.29YY297 pKa = 11.02AYY299 pKa = 8.86LTQMVYY305 pKa = 10.58DD306 pKa = 3.79QSQQRR311 pKa = 11.84LHH313 pKa = 5.98ICYY316 pKa = 9.75PSGNDD321 pKa = 3.57TLCTRR326 pKa = 11.84DD327 pKa = 3.54LFFDD331 pKa = 4.82FKK333 pKa = 11.12SNCWSLEE340 pKa = 3.67LLDD343 pKa = 4.65GYY345 pKa = 10.97SYY347 pKa = 10.89IGDD350 pKa = 3.66VFISSTPQLIYY361 pKa = 10.79EE362 pKa = 4.45DD363 pKa = 4.52ANMTFEE369 pKa = 6.26DD370 pKa = 3.9STDD373 pKa = 3.77DD374 pKa = 4.01FNLVYY379 pKa = 10.85DD380 pKa = 3.92KK381 pKa = 11.05FSNNRR386 pKa = 11.84LVYY389 pKa = 9.83FNGSIFQVGGEE400 pKa = 4.33SQVKK404 pKa = 9.31QATLTRR410 pKa = 11.84NLVAYY415 pKa = 6.56TTQDD419 pKa = 2.85TSGTTTVKK427 pKa = 10.64RR428 pKa = 11.84NLQLLITEE436 pKa = 5.16LWPKK440 pKa = 10.31LYY442 pKa = 10.54SGQVQFRR449 pKa = 11.84LGFSEE454 pKa = 4.69TPLGTITWSDD464 pKa = 3.08WVSTNGLDD472 pKa = 4.04RR473 pKa = 11.84LDD475 pKa = 4.21FFLSGLYY482 pKa = 10.25LHH484 pKa = 6.64LQIRR488 pKa = 11.84NEE490 pKa = 4.19GQDD493 pKa = 3.44FTLNGYY499 pKa = 7.56MLQAAPTGKK508 pKa = 9.8IAA510 pKa = 4.71
MM1 pKa = 7.25EE2 pKa = 5.54LEE4 pKa = 4.31IGSLHH9 pKa = 6.75SGGLVVGVNPLEE21 pKa = 4.16LAPNVFNDD29 pKa = 3.32VANVEE34 pKa = 4.55FTRR37 pKa = 11.84EE38 pKa = 3.82GVRR41 pKa = 11.84PLRR44 pKa = 11.84QPVTRR49 pKa = 11.84NLDD52 pKa = 3.5VPGSRR57 pKa = 11.84LVYY60 pKa = 9.96HH61 pKa = 7.07AVDD64 pKa = 3.3TMVNGGIRR72 pKa = 11.84VVLCFCEE79 pKa = 4.2DD80 pKa = 2.59KK81 pKa = 11.23VYY83 pKa = 11.09ALRR86 pKa = 11.84GSSYY90 pKa = 11.33LDD92 pKa = 3.22VTPAGMLSSKK102 pKa = 9.09EE103 pKa = 3.8WSVTEE108 pKa = 3.76FNGYY112 pKa = 8.47VVACNGKK119 pKa = 8.84NFPYY123 pKa = 10.62YY124 pKa = 10.71YY125 pKa = 10.54DD126 pKa = 3.71FADD129 pKa = 4.13ADD131 pKa = 4.22SVLKK135 pKa = 10.34ILPEE139 pKa = 3.86WPEE142 pKa = 3.52EE143 pKa = 3.8LVPQKK148 pKa = 10.39LSSMSGFLVFMGLVSDD164 pKa = 4.13GTFANQLIVWSDD176 pKa = 3.18AAEE179 pKa = 4.45LGSLPDD185 pKa = 3.53NYY187 pKa = 10.99NWADD191 pKa = 3.42PTSRR195 pKa = 11.84AGFYY199 pKa = 10.48SLPDD203 pKa = 3.49FEE205 pKa = 5.24EE206 pKa = 4.45FVAAVLLNKK215 pKa = 10.29SLIIYY220 pKa = 7.46RR221 pKa = 11.84TNSIYY226 pKa = 10.64EE227 pKa = 3.66MRR229 pKa = 11.84FIGGTLVFSIEE240 pKa = 3.69QRR242 pKa = 11.84FEE244 pKa = 3.87GKK246 pKa = 7.95TLISSKK252 pKa = 10.75AVAAKK257 pKa = 10.38GRR259 pKa = 11.84LHH261 pKa = 5.94YY262 pKa = 10.71CIGEE266 pKa = 4.08NEE268 pKa = 4.6FYY270 pKa = 11.41VHH272 pKa = 7.34DD273 pKa = 4.81GSSEE277 pKa = 3.96QVFDD281 pKa = 3.66VRR283 pKa = 11.84PVSDD287 pKa = 4.08YY288 pKa = 11.53YY289 pKa = 11.57FNTVNPDD296 pKa = 3.29YY297 pKa = 11.02AYY299 pKa = 8.86LTQMVYY305 pKa = 10.58DD306 pKa = 3.79QSQQRR311 pKa = 11.84LHH313 pKa = 5.98ICYY316 pKa = 9.75PSGNDD321 pKa = 3.57TLCTRR326 pKa = 11.84DD327 pKa = 3.54LFFDD331 pKa = 4.82FKK333 pKa = 11.12SNCWSLEE340 pKa = 3.67LLDD343 pKa = 4.65GYY345 pKa = 10.97SYY347 pKa = 10.89IGDD350 pKa = 3.66VFISSTPQLIYY361 pKa = 10.79EE362 pKa = 4.45DD363 pKa = 4.52ANMTFEE369 pKa = 6.26DD370 pKa = 3.9STDD373 pKa = 3.77DD374 pKa = 4.01FNLVYY379 pKa = 10.85DD380 pKa = 3.92KK381 pKa = 11.05FSNNRR386 pKa = 11.84LVYY389 pKa = 9.83FNGSIFQVGGEE400 pKa = 4.33SQVKK404 pKa = 9.31QATLTRR410 pKa = 11.84NLVAYY415 pKa = 6.56TTQDD419 pKa = 2.85TSGTTTVKK427 pKa = 10.64RR428 pKa = 11.84NLQLLITEE436 pKa = 5.16LWPKK440 pKa = 10.31LYY442 pKa = 10.54SGQVQFRR449 pKa = 11.84LGFSEE454 pKa = 4.69TPLGTITWSDD464 pKa = 3.08WVSTNGLDD472 pKa = 4.04RR473 pKa = 11.84LDD475 pKa = 4.21FFLSGLYY482 pKa = 10.25LHH484 pKa = 6.64LQIRR488 pKa = 11.84NEE490 pKa = 4.19GQDD493 pKa = 3.44FTLNGYY499 pKa = 7.56MLQAAPTGKK508 pKa = 9.8IAA510 pKa = 4.71
Molecular weight: 57.4 kDa
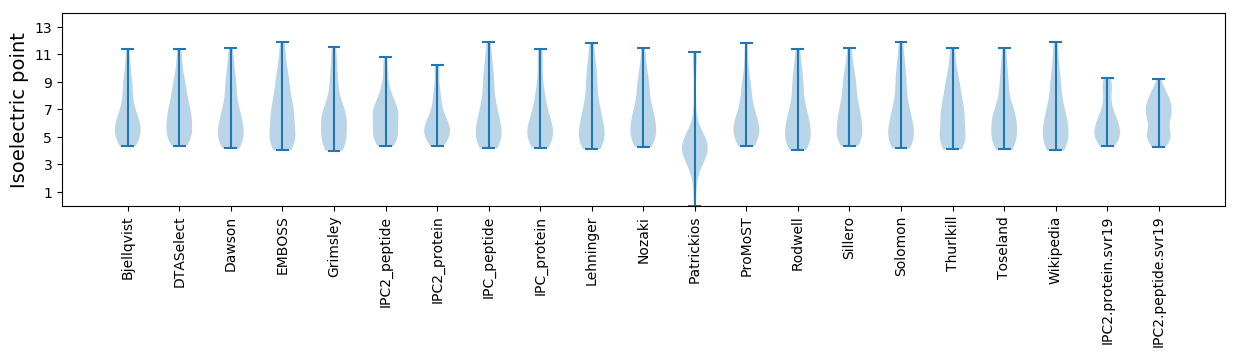
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WBI9|A0A5Q2WBI9_9CAUD Uncharacterized protein OS=Siphoviridae sp. ct7UA22 OX=2656718 PE=4 SV=1
MM1 pKa = 6.99VRR3 pKa = 11.84SFHH6 pKa = 6.98ALRR9 pKa = 11.84FSAWQRR15 pKa = 11.84EE16 pKa = 4.24KK17 pKa = 10.86PHH19 pKa = 6.89DD20 pKa = 4.66LGWYY24 pKa = 9.45IKK26 pKa = 10.43GAYY29 pKa = 9.16HH30 pKa = 5.42GQKK33 pKa = 9.7RR34 pKa = 11.84HH35 pKa = 4.86
MM1 pKa = 6.99VRR3 pKa = 11.84SFHH6 pKa = 6.98ALRR9 pKa = 11.84FSAWQRR15 pKa = 11.84EE16 pKa = 4.24KK17 pKa = 10.86PHH19 pKa = 6.89DD20 pKa = 4.66LGWYY24 pKa = 9.45IKK26 pKa = 10.43GAYY29 pKa = 9.16HH30 pKa = 5.42GQKK33 pKa = 9.7RR34 pKa = 11.84HH35 pKa = 4.86
Molecular weight: 4.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13272 |
29 |
1890 |
241.3 |
26.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.546 ± 0.514 | 1.07 ± 0.19 |
5.998 ± 0.233 | 6.005 ± 0.258 |
4.28 ± 0.238 | 6.299 ± 0.339 |
1.756 ± 0.182 | 4.83 ± 0.25 |
5.432 ± 0.327 | 9.411 ± 0.344 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.81 ± 0.171 | 4.204 ± 0.201 |
4.913 ± 0.258 | 4.671 ± 0.403 |
5.206 ± 0.266 | 6.201 ± 0.319 |
6.065 ± 0.33 | 6.608 ± 0.314 |
1.386 ± 0.105 | 3.308 ± 0.195 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
