
Bovine respiratory syncytial virus (strain A51908) (BRS)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Pneumoviridae; Orthopneumovirus; Bovine orthopneumovirus
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
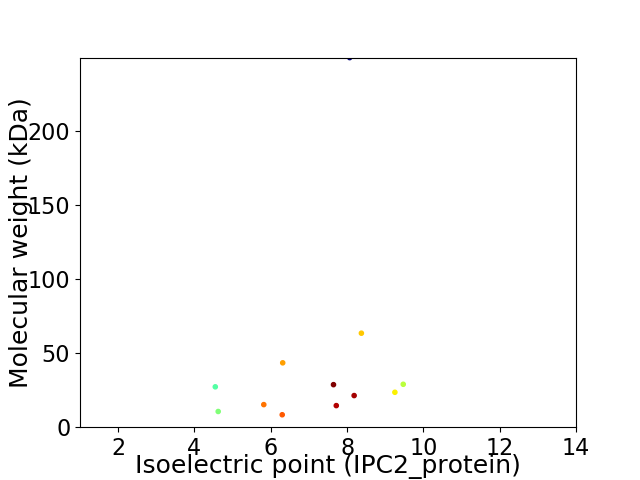
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
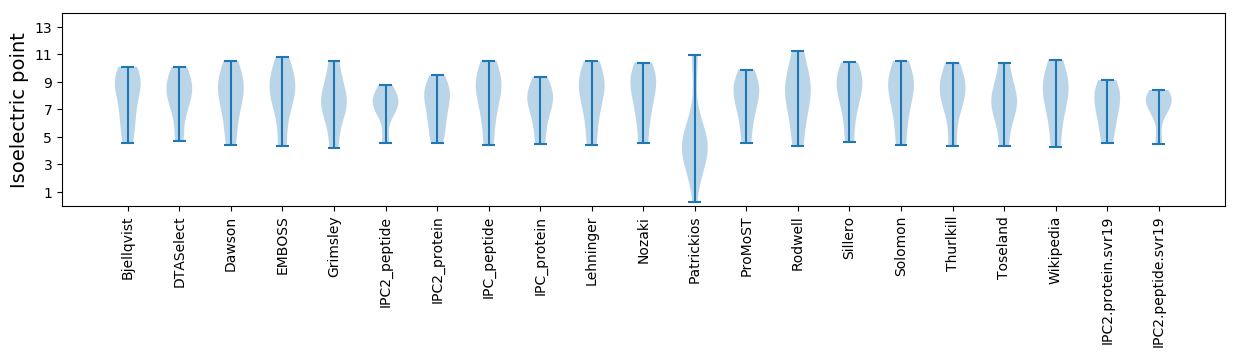
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P62648-2|GLYC-2_BRSVA Isoform of P62648 Isoform Secreted glycoprotein G of Major surface glycoprotein G OS=Bovine respiratory syncytial virus (strain A51908) OX=11247 GN=G PE=1 SV=1
MM1 pKa = 7.39NNSNIIIFPEE11 pKa = 4.59KK12 pKa = 9.62YY13 pKa = 8.38PCSISSLLIKK23 pKa = 10.5DD24 pKa = 3.88EE25 pKa = 4.14NDD27 pKa = 3.36VFVLSHH33 pKa = 6.36QNVLDD38 pKa = 4.04CLQFQYY44 pKa = 10.28PYY46 pKa = 11.7NMYY49 pKa = 10.05SQNHH53 pKa = 5.47MLDD56 pKa = 4.25DD57 pKa = 4.7IYY59 pKa = 9.89WTSQEE64 pKa = 4.6LIEE67 pKa = 4.95DD68 pKa = 3.77VLKK71 pKa = 10.59ILHH74 pKa = 6.63LSGISINKK82 pKa = 7.87YY83 pKa = 8.85VIYY86 pKa = 10.84VLVLL90 pKa = 3.28
MM1 pKa = 7.39NNSNIIIFPEE11 pKa = 4.59KK12 pKa = 9.62YY13 pKa = 8.38PCSISSLLIKK23 pKa = 10.5DD24 pKa = 3.88EE25 pKa = 4.14NDD27 pKa = 3.36VFVLSHH33 pKa = 6.36QNVLDD38 pKa = 4.04CLQFQYY44 pKa = 10.28PYY46 pKa = 11.7NMYY49 pKa = 10.05SQNHH53 pKa = 5.47MLDD56 pKa = 4.25DD57 pKa = 4.7IYY59 pKa = 9.89WTSQEE64 pKa = 4.6LIEE67 pKa = 4.95DD68 pKa = 3.77VLKK71 pKa = 10.59ILHH74 pKa = 6.63LSGISINKK82 pKa = 7.87YY83 pKa = 8.85VIYY86 pKa = 10.84VLVLL90 pKa = 3.28
Molecular weight: 10.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q65694|NS1_BRSVA Non-structural protein 1 OS=Bovine respiratory syncytial virus (strain A51908) OX=11247 GN=1C PE=2 SV=1
MM1 pKa = 7.56SNHH4 pKa = 4.32THH6 pKa = 5.99HH7 pKa = 7.36PKK9 pKa = 10.56FKK11 pKa = 8.79TLKK14 pKa = 9.66RR15 pKa = 11.84AWKK18 pKa = 8.2ASKK21 pKa = 10.57YY22 pKa = 10.19FIVGLSCLYY31 pKa = 10.63KK32 pKa = 10.77FNLKK36 pKa = 10.52SLVQTALTSLAMITLTSLVITAIIYY61 pKa = 10.23ISVGNAKK68 pKa = 10.16AKK70 pKa = 7.59PTSKK74 pKa = 9.21PTTQQTQQPQNHH86 pKa = 6.19TPLLPTEE93 pKa = 4.81HH94 pKa = 6.05NHH96 pKa = 6.54KK97 pKa = 9.28STHH100 pKa = 5.95TSTQSTTLSQPPNIDD115 pKa = 3.38TTSGTTYY122 pKa = 9.98GHH124 pKa = 7.56PINRR128 pKa = 11.84TQNRR132 pKa = 11.84KK133 pKa = 9.07IKK135 pKa = 10.34SQSTPLATRR144 pKa = 11.84KK145 pKa = 10.21LPINPLEE152 pKa = 4.39SNPPEE157 pKa = 3.73NHH159 pKa = 5.94QDD161 pKa = 3.42HH162 pKa = 6.87NNSQTLPHH170 pKa = 6.35VPCSTCEE177 pKa = 3.91GNPACSPLCQIGLEE191 pKa = 4.29RR192 pKa = 11.84APSRR196 pKa = 11.84APTITLKK203 pKa = 10.68KK204 pKa = 9.96APKK207 pKa = 9.76PKK209 pKa = 8.31TTKK212 pKa = 10.46KK213 pKa = 8.12PTKK216 pKa = 6.53TTIYY220 pKa = 10.39HH221 pKa = 5.41RR222 pKa = 11.84TSPEE226 pKa = 3.5AKK228 pKa = 10.14LQTKK232 pKa = 10.23KK233 pKa = 9.31NTATPQQGILSSPEE247 pKa = 3.74HH248 pKa = 5.42QTNQSTTQISQHH260 pKa = 4.92TSII263 pKa = 5.06
MM1 pKa = 7.56SNHH4 pKa = 4.32THH6 pKa = 5.99HH7 pKa = 7.36PKK9 pKa = 10.56FKK11 pKa = 8.79TLKK14 pKa = 9.66RR15 pKa = 11.84AWKK18 pKa = 8.2ASKK21 pKa = 10.57YY22 pKa = 10.19FIVGLSCLYY31 pKa = 10.63KK32 pKa = 10.77FNLKK36 pKa = 10.52SLVQTALTSLAMITLTSLVITAIIYY61 pKa = 10.23ISVGNAKK68 pKa = 10.16AKK70 pKa = 7.59PTSKK74 pKa = 9.21PTTQQTQQPQNHH86 pKa = 6.19TPLLPTEE93 pKa = 4.81HH94 pKa = 6.05NHH96 pKa = 6.54KK97 pKa = 9.28STHH100 pKa = 5.95TSTQSTTLSQPPNIDD115 pKa = 3.38TTSGTTYY122 pKa = 9.98GHH124 pKa = 7.56PINRR128 pKa = 11.84TQNRR132 pKa = 11.84KK133 pKa = 9.07IKK135 pKa = 10.34SQSTPLATRR144 pKa = 11.84KK145 pKa = 10.21LPINPLEE152 pKa = 4.39SNPPEE157 pKa = 3.73NHH159 pKa = 5.94QDD161 pKa = 3.42HH162 pKa = 6.87NNSQTLPHH170 pKa = 6.35VPCSTCEE177 pKa = 3.91GNPACSPLCQIGLEE191 pKa = 4.29RR192 pKa = 11.84APSRR196 pKa = 11.84APTITLKK203 pKa = 10.68KK204 pKa = 9.96APKK207 pKa = 9.76PKK209 pKa = 8.31TTKK212 pKa = 10.46KK213 pKa = 8.12PTKK216 pKa = 6.53TTIYY220 pKa = 10.39HH221 pKa = 5.41RR222 pKa = 11.84TSPEE226 pKa = 3.5AKK228 pKa = 10.14LQTKK232 pKa = 10.23KK233 pKa = 9.31NTATPQQGILSSPEE247 pKa = 3.74HH248 pKa = 5.42QTNQSTTQISQHH260 pKa = 4.92TSII263 pKa = 5.06
Molecular weight: 28.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4710 |
73 |
2162 |
392.5 |
44.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.268 ± 0.663 | 1.847 ± 0.258 |
4.756 ± 0.476 | 4.968 ± 0.742 |
3.397 ± 0.449 | 4.034 ± 0.504 |
2.76 ± 0.387 | 8.896 ± 0.488 |
7.558 ± 0.275 | 10.849 ± 1.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.251 ± 0.269 | 7.197 ± 0.285 |
4.013 ± 0.718 | 3.461 ± 0.515 |
3.652 ± 0.256 | 8.174 ± 0.553 |
7.877 ± 1.146 | 5.074 ± 0.685 |
0.786 ± 0.224 | 4.183 ± 0.572 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
