
Labrenzia sp. OB1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Stappiaceae; Labrenzia; unclassified Labrenzia
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
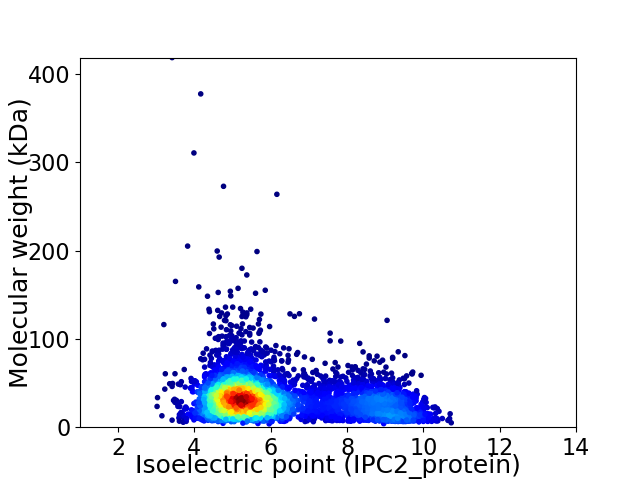
Virtual 2D-PAGE plot for 5016 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
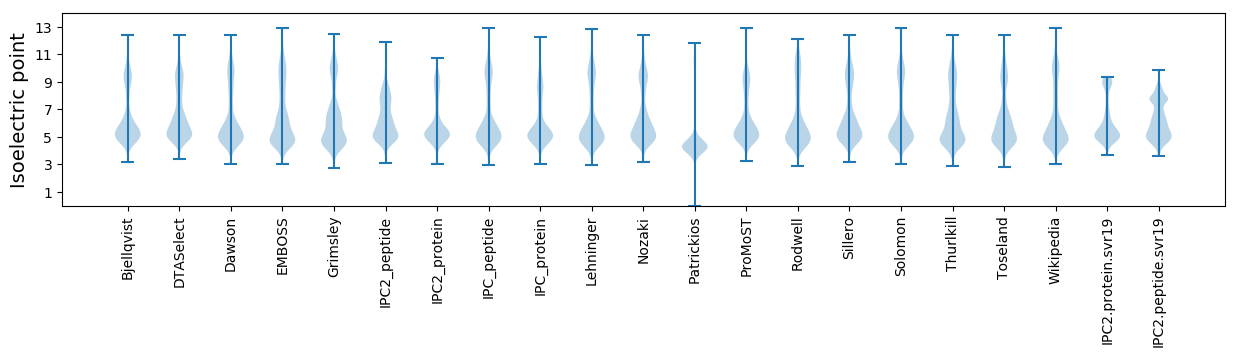
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161YYJ8|A0A161YYJ8_9RHOB Uncharacterized protein OS=Labrenzia sp. OB1 OX=1561204 GN=OA90_15465 PE=4 SV=1
MM1 pKa = 7.33QSTGAQGQDD10 pKa = 2.53TGTAGNGGEE19 pKa = 3.56IRR21 pKa = 11.84IYY23 pKa = 9.9NDD25 pKa = 2.79SYY27 pKa = 11.64INYY30 pKa = 9.09QVGNTSSLAVGMNARR45 pKa = 11.84SLGGAGDD52 pKa = 3.62QDD54 pKa = 4.57NRR56 pKa = 11.84DD57 pKa = 3.48NDD59 pKa = 4.42SNGGTGGNGGSLYY72 pKa = 9.39FTSDD76 pKa = 3.62GIAGYY81 pKa = 10.4DD82 pKa = 3.28VDD84 pKa = 4.43GGSNAGFVIYY94 pKa = 10.47LGEE97 pKa = 4.33SYY99 pKa = 11.26GGMGGKK105 pKa = 9.87QNDD108 pKa = 4.1SVLSDD113 pKa = 3.23QRR115 pKa = 11.84GGQGGSGGAITIVNNSLASFDD136 pKa = 3.9IGSSSSRR143 pKa = 11.84FTGMGSGGGFFAKK156 pKa = 10.24SVGGQGGFNNGNAGAGGAVSITGDD180 pKa = 3.95LDD182 pKa = 3.66GSIYY186 pKa = 10.27WDD188 pKa = 3.49VTAGGGTGLFGLSGLSQGGDD208 pKa = 3.17GTTSTDD214 pKa = 3.14NSDD217 pKa = 3.28NGGNGGNGGTVTISNGEE234 pKa = 4.07GLVLDD239 pKa = 3.79VLGTVGEE246 pKa = 4.38ISAVLAGKK254 pKa = 9.72SAGGNGGTGPAEE266 pKa = 3.84DD267 pKa = 4.71HH268 pKa = 6.65YY269 pKa = 11.19GGSGGTAGAVSLSLNNDD286 pKa = 2.71GDD288 pKa = 4.18LAASGDD294 pKa = 4.08YY295 pKa = 10.9VYY297 pKa = 11.14GILGQSIGGEE307 pKa = 4.21GGDD310 pKa = 4.34GGDD313 pKa = 3.64GTALAGTGGGGGFGGNAGNVTVSVDD338 pKa = 2.49IWAGSVSTNGDD349 pKa = 3.52YY350 pKa = 11.3APAIAAHH357 pKa = 6.36SIGGGGGTGGDD368 pKa = 3.7FVSVLGGQAGNGGNGGNAGNVLVSLDD394 pKa = 3.77AQGLTTSGDD403 pKa = 3.26HH404 pKa = 6.49SFGIVAQSIAGSGGTGGIDD423 pKa = 3.04TSTLVGLGGDD433 pKa = 3.91GAGGGSVGTVTINSSTPITTQGFNALGIIGQSIGGGGGAAGSASGLVSVGGNAQGSSTSDD493 pKa = 2.84GGTVGISNTEE503 pKa = 3.98SVVTKK508 pKa = 10.31GEE510 pKa = 3.92SSIAILAQSIGGGGGSGGDD529 pKa = 3.21ATGIAGVGGQGAAGGDD545 pKa = 3.6GGIIAIPNIGTITTYY560 pKa = 11.27DD561 pKa = 3.65DD562 pKa = 3.88SSPGAVFQSIGGGGGNGGSTLTLSTIASIGIGGSASGGGDD602 pKa = 3.24GGLLCIDD609 pKa = 3.96NSAVCGANNSSGTFVPYY626 pKa = 10.63LSSLASPEE634 pKa = 4.14DD635 pKa = 4.1DD636 pKa = 3.07GDD638 pKa = 4.1ARR640 pKa = 11.84VTTIGDD646 pKa = 4.15FAPGVVAQSVGGGGGNGGSDD666 pKa = 3.45KK667 pKa = 11.08NYY669 pKa = 10.06SALSFFALQMGGSAGAGGAGGTVRR693 pKa = 11.84FVQDD697 pKa = 3.16EE698 pKa = 4.46FAVNTSGAQSPGLIAQSIGGGGGTGGSASYY728 pKa = 11.25YY729 pKa = 10.41DD730 pKa = 3.41ATIGFNAAFIVGGSGGAGGSGGTLNLDD757 pKa = 3.28LTDD760 pKa = 4.6AYY762 pKa = 9.92IATGLITNSTTQSAPNNSVGILAQSIGGGGGSGGAVSASDD802 pKa = 3.52FVVAAPTGTGVPVAFNYY819 pKa = 9.08QAAVGGNGANAGNGGSVTVSFDD841 pKa = 3.37NDD843 pKa = 3.41SGLTTIGDD851 pKa = 3.84AAHH854 pKa = 7.04GIVAQSIGGGGGNGGDD870 pKa = 3.41ASVLSTTLGDD880 pKa = 3.51KK881 pKa = 10.2DD882 pKa = 4.11TVEE885 pKa = 4.32VTAGVAFGGGAGNIMSDD902 pKa = 3.83FNPGYY907 pKa = 10.61SYY909 pKa = 7.69TTSGGGAGGSVSVTLGTYY927 pKa = 10.53GEE929 pKa = 4.29YY930 pKa = 10.67TNASGTGATRR940 pKa = 11.84LTTLAPDD947 pKa = 3.62GTMLTVGDD955 pKa = 4.21GAHH958 pKa = 6.48GVLAQSIGGGGGNGGVGNSNAYY980 pKa = 9.37AQGGVASLKK989 pKa = 10.69ADD991 pKa = 3.21ISLGGQGGSGGNGNSVTVHH1010 pKa = 5.87NFAGYY1015 pKa = 7.59TVQTQGSGARR1025 pKa = 11.84GIVAQSIGGGGGNSQGGTLYY1045 pKa = 10.9LAANAGGFSGRR1056 pKa = 11.84LNVGVGATGGSGGEE1070 pKa = 4.13GGSVYY1075 pKa = 10.52VEE1077 pKa = 4.16EE1078 pKa = 4.65YY1079 pKa = 11.07GVTEE1083 pKa = 4.3TFGGDD1088 pKa = 2.87ADD1090 pKa = 4.9GIVLQSIGGGGGIGGSLGSDD1110 pKa = 3.07ASSHH1114 pKa = 6.8KK1115 pKa = 10.69ILDD1118 pKa = 3.71QIGKK1122 pKa = 9.41IKK1124 pKa = 11.05NNLSRR1129 pKa = 11.84LTDD1132 pKa = 3.47AGSSYY1137 pKa = 11.13EE1138 pKa = 3.92LTVDD1142 pKa = 3.58VGGKK1146 pKa = 9.59GGNGGDD1152 pKa = 3.64GGTVKK1157 pKa = 10.58YY1158 pKa = 9.89YY1159 pKa = 9.7QAGRR1163 pKa = 11.84VATYY1167 pKa = 10.83GDD1169 pKa = 3.43WADD1172 pKa = 3.61GVVLQSIGGGGGQGGSSVATGGDD1195 pKa = 3.09VTANIEE1201 pKa = 4.18VSVGGTGGSGGVGGEE1216 pKa = 3.66AAIWVSGQGSNLVEE1230 pKa = 3.91THH1232 pKa = 6.75GYY1234 pKa = 8.56GAAGLLMQSIGGGGGQGGDD1253 pKa = 3.68GSDD1256 pKa = 3.59SQTGTMAIGGDD1267 pKa = 3.25WGGSGGAGNDD1277 pKa = 3.45GGSVYY1282 pKa = 10.13TYY1284 pKa = 10.17NDD1286 pKa = 2.9SDD1288 pKa = 3.99IFINTYY1294 pKa = 10.47GSDD1297 pKa = 3.53AAGLIAQAIGGGGGTGGVGSSVEE1320 pKa = 3.77ATRR1323 pKa = 11.84ADD1325 pKa = 3.35GHH1327 pKa = 5.74EE1328 pKa = 4.34LSVSVGGSGGVAGDD1342 pKa = 3.52GGTINLQADD1351 pKa = 3.94TSIVTLGNRR1360 pKa = 11.84SHH1362 pKa = 6.57GVLLQSIGGGGGLGVTGSVGDD1383 pKa = 4.71DD1384 pKa = 3.16LDD1386 pKa = 4.06LAIGGTGGAAGDD1398 pKa = 4.48GNTVTYY1404 pKa = 10.64SLTGGTGVATGGIGAHH1420 pKa = 5.56GVIAQSIGGGGGLGGDD1436 pKa = 3.39ATGGVLSVLTSGDD1449 pKa = 3.56NASGGASGNGGTITLDD1465 pKa = 2.76IAAPIAVTGQDD1476 pKa = 3.39AIGVVAQSIGGGGGLGGSGTGAFAGVTSSDD1506 pKa = 3.29GSGTGGDD1513 pKa = 3.55VTLTISDD1520 pKa = 4.08KK1521 pKa = 10.96VSSTGTRR1528 pKa = 11.84GMGVFAQSQGPDD1540 pKa = 3.1GNEE1543 pKa = 3.22QVMITTNSGGQIIGGYY1559 pKa = 8.81SDD1561 pKa = 3.68SSSAVMVAGGNGNSLVLNGDD1581 pKa = 3.85SVVEE1585 pKa = 4.17SGSYY1589 pKa = 7.51VTGNDD1594 pKa = 3.11YY1595 pKa = 11.53DD1596 pKa = 4.33DD1597 pKa = 4.67NYY1599 pKa = 11.19AVRR1602 pKa = 11.84YY1603 pKa = 9.17SGSGTTAEE1611 pKa = 4.21GAILDD1616 pKa = 3.85VTVNDD1621 pKa = 3.99TASLHH1626 pKa = 6.53GDD1628 pKa = 3.51VLLEE1632 pKa = 4.23NADD1635 pKa = 3.64GNTAGTVTNNSSNTLAGGYY1654 pKa = 9.81FYY1656 pKa = 11.04GAHH1659 pKa = 6.15IVNNGRR1665 pKa = 11.84FVLNRR1670 pKa = 11.84EE1671 pKa = 4.06HH1672 pKa = 7.35DD1673 pKa = 3.45TGTTVLTGNFIQSDD1687 pKa = 3.75AGKK1690 pKa = 10.04LVADD1694 pKa = 4.21IDD1696 pKa = 4.43LNNYY1700 pKa = 9.74GSDD1703 pKa = 3.44LFDD1706 pKa = 3.28IRR1708 pKa = 11.84GDD1710 pKa = 3.46ASLDD1714 pKa = 3.3GTLGVDD1720 pKa = 5.08AISLLPSRR1728 pKa = 11.84RR1729 pKa = 11.84TRR1731 pKa = 11.84FLTVGGTLSGSLTPQDD1747 pKa = 4.14SLIFDD1752 pKa = 3.66YY1753 pKa = 11.05AINKK1757 pKa = 9.57IGTDD1761 pKa = 3.04HH1762 pKa = 6.22YY1763 pKa = 11.22LSVEE1767 pKa = 4.23GADD1770 pKa = 3.59FTPVDD1775 pKa = 3.9KK1776 pKa = 10.1TQMNDD1781 pKa = 3.14DD1782 pKa = 4.17QIRR1785 pKa = 11.84LAKK1788 pKa = 10.14NLQSIWNLGGNNAFGSLFGAIANEE1812 pKa = 3.75VDD1814 pKa = 3.31NAGRR1818 pKa = 11.84DD1819 pKa = 3.63YY1820 pKa = 11.71GDD1822 pKa = 3.52TLDD1825 pKa = 3.81EE1826 pKa = 4.34LRR1828 pKa = 11.84PRR1830 pKa = 11.84AALAPAVEE1838 pKa = 4.62QSFEE1842 pKa = 3.83MLAFGNSLMSCPVFEE1857 pKa = 5.57ADD1859 pKa = 4.34NAFLSEE1865 pKa = 4.92GSCGWARR1872 pKa = 11.84LTAATGHH1879 pKa = 5.6RR1880 pKa = 11.84QSMGTAPSYY1889 pKa = 10.93NADD1892 pKa = 2.97AVTYY1896 pKa = 10.33TMGAQKK1902 pKa = 10.25EE1903 pKa = 4.22IAPGWFVGTAAAFQQSWISQASNSVSGEE1931 pKa = 4.18GIGGFAGVSLKK1942 pKa = 10.92HH1943 pKa = 6.57EE1944 pKa = 4.55IGDD1947 pKa = 3.71WTLSGAVSGGYY1958 pKa = 8.95GQYY1961 pKa = 11.18DD1962 pKa = 4.02LDD1964 pKa = 3.94RR1965 pKa = 11.84RR1966 pKa = 11.84ISITNVSEE1974 pKa = 4.48TVSSSPNVYY1983 pKa = 10.27NGAIRR1988 pKa = 11.84ARR1990 pKa = 11.84IARR1993 pKa = 11.84TFAATDD1999 pKa = 4.45FYY2001 pKa = 11.58AKK2003 pKa = 10.24PYY2005 pKa = 10.08VDD2007 pKa = 4.96LDD2009 pKa = 3.57AVYY2012 pKa = 11.0SRR2014 pKa = 11.84VGEE2017 pKa = 4.09YY2018 pKa = 10.65KK2019 pKa = 8.95EE2020 pKa = 4.02TGVYY2024 pKa = 10.46GLEE2027 pKa = 4.66FDD2029 pKa = 5.73EE2030 pKa = 4.24TDD2032 pKa = 2.71QWNFAVSPSLEE2043 pKa = 4.07LGSWYY2048 pKa = 9.61TYY2050 pKa = 11.15DD2051 pKa = 3.15SGASLRR2057 pKa = 11.84FFGRR2061 pKa = 11.84VGLTVLAKK2069 pKa = 10.57DD2070 pKa = 3.76DD2071 pKa = 4.1WQTSAQFTSAPAGTGKK2087 pKa = 8.61FTTTLDD2093 pKa = 3.67HH2094 pKa = 7.02EE2095 pKa = 5.14DD2096 pKa = 3.37VFASIGAGLQLTSIDD2111 pKa = 3.95GFDD2114 pKa = 4.79LRR2116 pKa = 11.84LEE2118 pKa = 3.96YY2119 pKa = 10.54DD2120 pKa = 3.39GRR2122 pKa = 11.84FSEE2125 pKa = 5.66KK2126 pKa = 9.5MATNSGSFRR2135 pKa = 11.84VSIPFF2140 pKa = 3.65
MM1 pKa = 7.33QSTGAQGQDD10 pKa = 2.53TGTAGNGGEE19 pKa = 3.56IRR21 pKa = 11.84IYY23 pKa = 9.9NDD25 pKa = 2.79SYY27 pKa = 11.64INYY30 pKa = 9.09QVGNTSSLAVGMNARR45 pKa = 11.84SLGGAGDD52 pKa = 3.62QDD54 pKa = 4.57NRR56 pKa = 11.84DD57 pKa = 3.48NDD59 pKa = 4.42SNGGTGGNGGSLYY72 pKa = 9.39FTSDD76 pKa = 3.62GIAGYY81 pKa = 10.4DD82 pKa = 3.28VDD84 pKa = 4.43GGSNAGFVIYY94 pKa = 10.47LGEE97 pKa = 4.33SYY99 pKa = 11.26GGMGGKK105 pKa = 9.87QNDD108 pKa = 4.1SVLSDD113 pKa = 3.23QRR115 pKa = 11.84GGQGGSGGAITIVNNSLASFDD136 pKa = 3.9IGSSSSRR143 pKa = 11.84FTGMGSGGGFFAKK156 pKa = 10.24SVGGQGGFNNGNAGAGGAVSITGDD180 pKa = 3.95LDD182 pKa = 3.66GSIYY186 pKa = 10.27WDD188 pKa = 3.49VTAGGGTGLFGLSGLSQGGDD208 pKa = 3.17GTTSTDD214 pKa = 3.14NSDD217 pKa = 3.28NGGNGGNGGTVTISNGEE234 pKa = 4.07GLVLDD239 pKa = 3.79VLGTVGEE246 pKa = 4.38ISAVLAGKK254 pKa = 9.72SAGGNGGTGPAEE266 pKa = 3.84DD267 pKa = 4.71HH268 pKa = 6.65YY269 pKa = 11.19GGSGGTAGAVSLSLNNDD286 pKa = 2.71GDD288 pKa = 4.18LAASGDD294 pKa = 4.08YY295 pKa = 10.9VYY297 pKa = 11.14GILGQSIGGEE307 pKa = 4.21GGDD310 pKa = 4.34GGDD313 pKa = 3.64GTALAGTGGGGGFGGNAGNVTVSVDD338 pKa = 2.49IWAGSVSTNGDD349 pKa = 3.52YY350 pKa = 11.3APAIAAHH357 pKa = 6.36SIGGGGGTGGDD368 pKa = 3.7FVSVLGGQAGNGGNGGNAGNVLVSLDD394 pKa = 3.77AQGLTTSGDD403 pKa = 3.26HH404 pKa = 6.49SFGIVAQSIAGSGGTGGIDD423 pKa = 3.04TSTLVGLGGDD433 pKa = 3.91GAGGGSVGTVTINSSTPITTQGFNALGIIGQSIGGGGGAAGSASGLVSVGGNAQGSSTSDD493 pKa = 2.84GGTVGISNTEE503 pKa = 3.98SVVTKK508 pKa = 10.31GEE510 pKa = 3.92SSIAILAQSIGGGGGSGGDD529 pKa = 3.21ATGIAGVGGQGAAGGDD545 pKa = 3.6GGIIAIPNIGTITTYY560 pKa = 11.27DD561 pKa = 3.65DD562 pKa = 3.88SSPGAVFQSIGGGGGNGGSTLTLSTIASIGIGGSASGGGDD602 pKa = 3.24GGLLCIDD609 pKa = 3.96NSAVCGANNSSGTFVPYY626 pKa = 10.63LSSLASPEE634 pKa = 4.14DD635 pKa = 4.1DD636 pKa = 3.07GDD638 pKa = 4.1ARR640 pKa = 11.84VTTIGDD646 pKa = 4.15FAPGVVAQSVGGGGGNGGSDD666 pKa = 3.45KK667 pKa = 11.08NYY669 pKa = 10.06SALSFFALQMGGSAGAGGAGGTVRR693 pKa = 11.84FVQDD697 pKa = 3.16EE698 pKa = 4.46FAVNTSGAQSPGLIAQSIGGGGGTGGSASYY728 pKa = 11.25YY729 pKa = 10.41DD730 pKa = 3.41ATIGFNAAFIVGGSGGAGGSGGTLNLDD757 pKa = 3.28LTDD760 pKa = 4.6AYY762 pKa = 9.92IATGLITNSTTQSAPNNSVGILAQSIGGGGGSGGAVSASDD802 pKa = 3.52FVVAAPTGTGVPVAFNYY819 pKa = 9.08QAAVGGNGANAGNGGSVTVSFDD841 pKa = 3.37NDD843 pKa = 3.41SGLTTIGDD851 pKa = 3.84AAHH854 pKa = 7.04GIVAQSIGGGGGNGGDD870 pKa = 3.41ASVLSTTLGDD880 pKa = 3.51KK881 pKa = 10.2DD882 pKa = 4.11TVEE885 pKa = 4.32VTAGVAFGGGAGNIMSDD902 pKa = 3.83FNPGYY907 pKa = 10.61SYY909 pKa = 7.69TTSGGGAGGSVSVTLGTYY927 pKa = 10.53GEE929 pKa = 4.29YY930 pKa = 10.67TNASGTGATRR940 pKa = 11.84LTTLAPDD947 pKa = 3.62GTMLTVGDD955 pKa = 4.21GAHH958 pKa = 6.48GVLAQSIGGGGGNGGVGNSNAYY980 pKa = 9.37AQGGVASLKK989 pKa = 10.69ADD991 pKa = 3.21ISLGGQGGSGGNGNSVTVHH1010 pKa = 5.87NFAGYY1015 pKa = 7.59TVQTQGSGARR1025 pKa = 11.84GIVAQSIGGGGGNSQGGTLYY1045 pKa = 10.9LAANAGGFSGRR1056 pKa = 11.84LNVGVGATGGSGGEE1070 pKa = 4.13GGSVYY1075 pKa = 10.52VEE1077 pKa = 4.16EE1078 pKa = 4.65YY1079 pKa = 11.07GVTEE1083 pKa = 4.3TFGGDD1088 pKa = 2.87ADD1090 pKa = 4.9GIVLQSIGGGGGIGGSLGSDD1110 pKa = 3.07ASSHH1114 pKa = 6.8KK1115 pKa = 10.69ILDD1118 pKa = 3.71QIGKK1122 pKa = 9.41IKK1124 pKa = 11.05NNLSRR1129 pKa = 11.84LTDD1132 pKa = 3.47AGSSYY1137 pKa = 11.13EE1138 pKa = 3.92LTVDD1142 pKa = 3.58VGGKK1146 pKa = 9.59GGNGGDD1152 pKa = 3.64GGTVKK1157 pKa = 10.58YY1158 pKa = 9.89YY1159 pKa = 9.7QAGRR1163 pKa = 11.84VATYY1167 pKa = 10.83GDD1169 pKa = 3.43WADD1172 pKa = 3.61GVVLQSIGGGGGQGGSSVATGGDD1195 pKa = 3.09VTANIEE1201 pKa = 4.18VSVGGTGGSGGVGGEE1216 pKa = 3.66AAIWVSGQGSNLVEE1230 pKa = 3.91THH1232 pKa = 6.75GYY1234 pKa = 8.56GAAGLLMQSIGGGGGQGGDD1253 pKa = 3.68GSDD1256 pKa = 3.59SQTGTMAIGGDD1267 pKa = 3.25WGGSGGAGNDD1277 pKa = 3.45GGSVYY1282 pKa = 10.13TYY1284 pKa = 10.17NDD1286 pKa = 2.9SDD1288 pKa = 3.99IFINTYY1294 pKa = 10.47GSDD1297 pKa = 3.53AAGLIAQAIGGGGGTGGVGSSVEE1320 pKa = 3.77ATRR1323 pKa = 11.84ADD1325 pKa = 3.35GHH1327 pKa = 5.74EE1328 pKa = 4.34LSVSVGGSGGVAGDD1342 pKa = 3.52GGTINLQADD1351 pKa = 3.94TSIVTLGNRR1360 pKa = 11.84SHH1362 pKa = 6.57GVLLQSIGGGGGLGVTGSVGDD1383 pKa = 4.71DD1384 pKa = 3.16LDD1386 pKa = 4.06LAIGGTGGAAGDD1398 pKa = 4.48GNTVTYY1404 pKa = 10.64SLTGGTGVATGGIGAHH1420 pKa = 5.56GVIAQSIGGGGGLGGDD1436 pKa = 3.39ATGGVLSVLTSGDD1449 pKa = 3.56NASGGASGNGGTITLDD1465 pKa = 2.76IAAPIAVTGQDD1476 pKa = 3.39AIGVVAQSIGGGGGLGGSGTGAFAGVTSSDD1506 pKa = 3.29GSGTGGDD1513 pKa = 3.55VTLTISDD1520 pKa = 4.08KK1521 pKa = 10.96VSSTGTRR1528 pKa = 11.84GMGVFAQSQGPDD1540 pKa = 3.1GNEE1543 pKa = 3.22QVMITTNSGGQIIGGYY1559 pKa = 8.81SDD1561 pKa = 3.68SSSAVMVAGGNGNSLVLNGDD1581 pKa = 3.85SVVEE1585 pKa = 4.17SGSYY1589 pKa = 7.51VTGNDD1594 pKa = 3.11YY1595 pKa = 11.53DD1596 pKa = 4.33DD1597 pKa = 4.67NYY1599 pKa = 11.19AVRR1602 pKa = 11.84YY1603 pKa = 9.17SGSGTTAEE1611 pKa = 4.21GAILDD1616 pKa = 3.85VTVNDD1621 pKa = 3.99TASLHH1626 pKa = 6.53GDD1628 pKa = 3.51VLLEE1632 pKa = 4.23NADD1635 pKa = 3.64GNTAGTVTNNSSNTLAGGYY1654 pKa = 9.81FYY1656 pKa = 11.04GAHH1659 pKa = 6.15IVNNGRR1665 pKa = 11.84FVLNRR1670 pKa = 11.84EE1671 pKa = 4.06HH1672 pKa = 7.35DD1673 pKa = 3.45TGTTVLTGNFIQSDD1687 pKa = 3.75AGKK1690 pKa = 10.04LVADD1694 pKa = 4.21IDD1696 pKa = 4.43LNNYY1700 pKa = 9.74GSDD1703 pKa = 3.44LFDD1706 pKa = 3.28IRR1708 pKa = 11.84GDD1710 pKa = 3.46ASLDD1714 pKa = 3.3GTLGVDD1720 pKa = 5.08AISLLPSRR1728 pKa = 11.84RR1729 pKa = 11.84TRR1731 pKa = 11.84FLTVGGTLSGSLTPQDD1747 pKa = 4.14SLIFDD1752 pKa = 3.66YY1753 pKa = 11.05AINKK1757 pKa = 9.57IGTDD1761 pKa = 3.04HH1762 pKa = 6.22YY1763 pKa = 11.22LSVEE1767 pKa = 4.23GADD1770 pKa = 3.59FTPVDD1775 pKa = 3.9KK1776 pKa = 10.1TQMNDD1781 pKa = 3.14DD1782 pKa = 4.17QIRR1785 pKa = 11.84LAKK1788 pKa = 10.14NLQSIWNLGGNNAFGSLFGAIANEE1812 pKa = 3.75VDD1814 pKa = 3.31NAGRR1818 pKa = 11.84DD1819 pKa = 3.63YY1820 pKa = 11.71GDD1822 pKa = 3.52TLDD1825 pKa = 3.81EE1826 pKa = 4.34LRR1828 pKa = 11.84PRR1830 pKa = 11.84AALAPAVEE1838 pKa = 4.62QSFEE1842 pKa = 3.83MLAFGNSLMSCPVFEE1857 pKa = 5.57ADD1859 pKa = 4.34NAFLSEE1865 pKa = 4.92GSCGWARR1872 pKa = 11.84LTAATGHH1879 pKa = 5.6RR1880 pKa = 11.84QSMGTAPSYY1889 pKa = 10.93NADD1892 pKa = 2.97AVTYY1896 pKa = 10.33TMGAQKK1902 pKa = 10.25EE1903 pKa = 4.22IAPGWFVGTAAAFQQSWISQASNSVSGEE1931 pKa = 4.18GIGGFAGVSLKK1942 pKa = 10.92HH1943 pKa = 6.57EE1944 pKa = 4.55IGDD1947 pKa = 3.71WTLSGAVSGGYY1958 pKa = 8.95GQYY1961 pKa = 11.18DD1962 pKa = 4.02LDD1964 pKa = 3.94RR1965 pKa = 11.84RR1966 pKa = 11.84ISITNVSEE1974 pKa = 4.48TVSSSPNVYY1983 pKa = 10.27NGAIRR1988 pKa = 11.84ARR1990 pKa = 11.84IARR1993 pKa = 11.84TFAATDD1999 pKa = 4.45FYY2001 pKa = 11.58AKK2003 pKa = 10.24PYY2005 pKa = 10.08VDD2007 pKa = 4.96LDD2009 pKa = 3.57AVYY2012 pKa = 11.0SRR2014 pKa = 11.84VGEE2017 pKa = 4.09YY2018 pKa = 10.65KK2019 pKa = 8.95EE2020 pKa = 4.02TGVYY2024 pKa = 10.46GLEE2027 pKa = 4.66FDD2029 pKa = 5.73EE2030 pKa = 4.24TDD2032 pKa = 2.71QWNFAVSPSLEE2043 pKa = 4.07LGSWYY2048 pKa = 9.61TYY2050 pKa = 11.15DD2051 pKa = 3.15SGASLRR2057 pKa = 11.84FFGRR2061 pKa = 11.84VGLTVLAKK2069 pKa = 10.57DD2070 pKa = 3.76DD2071 pKa = 4.1WQTSAQFTSAPAGTGKK2087 pKa = 8.61FTTTLDD2093 pKa = 3.67HH2094 pKa = 7.02EE2095 pKa = 5.14DD2096 pKa = 3.37VFASIGAGLQLTSIDD2111 pKa = 3.95GFDD2114 pKa = 4.79LRR2116 pKa = 11.84LEE2118 pKa = 3.96YY2119 pKa = 10.54DD2120 pKa = 3.39GRR2122 pKa = 11.84FSEE2125 pKa = 5.66KK2126 pKa = 9.5MATNSGSFRR2135 pKa = 11.84VSIPFF2140 pKa = 3.65
Molecular weight: 205.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A163U730|A0A163U730_9RHOB Uncharacterized protein OS=Labrenzia sp. OB1 OX=1561204 GN=OA90_21190 PE=4 SV=1
MM1 pKa = 7.56ALKK4 pKa = 10.08KK5 pKa = 9.9ICYY8 pKa = 9.65RR9 pKa = 11.84LEE11 pKa = 3.99NRR13 pKa = 11.84NIACEE18 pKa = 3.83FGVFGKK24 pKa = 10.0DD25 pKa = 3.01CRR27 pKa = 11.84QVAEE31 pKa = 4.2KK32 pKa = 10.08AQRR35 pKa = 11.84RR36 pKa = 11.84HH37 pKa = 4.2RR38 pKa = 11.84HH39 pKa = 5.64RR40 pKa = 11.84YY41 pKa = 7.74GPSPIMDD48 pKa = 4.39GNRR51 pKa = 11.84EE52 pKa = 4.0RR53 pKa = 11.84APGDD57 pKa = 3.51LAEE60 pKa = 4.63KK61 pKa = 10.52DD62 pKa = 3.33IVRR65 pKa = 11.84EE66 pKa = 3.63RR67 pKa = 11.84LIRR70 pKa = 11.84LFVRR74 pKa = 11.84RR75 pKa = 11.84LEE77 pKa = 4.15VRR79 pKa = 3.29
MM1 pKa = 7.56ALKK4 pKa = 10.08KK5 pKa = 9.9ICYY8 pKa = 9.65RR9 pKa = 11.84LEE11 pKa = 3.99NRR13 pKa = 11.84NIACEE18 pKa = 3.83FGVFGKK24 pKa = 10.0DD25 pKa = 3.01CRR27 pKa = 11.84QVAEE31 pKa = 4.2KK32 pKa = 10.08AQRR35 pKa = 11.84RR36 pKa = 11.84HH37 pKa = 4.2RR38 pKa = 11.84HH39 pKa = 5.64RR40 pKa = 11.84YY41 pKa = 7.74GPSPIMDD48 pKa = 4.39GNRR51 pKa = 11.84EE52 pKa = 4.0RR53 pKa = 11.84APGDD57 pKa = 3.51LAEE60 pKa = 4.63KK61 pKa = 10.52DD62 pKa = 3.33IVRR65 pKa = 11.84EE66 pKa = 3.63RR67 pKa = 11.84LIRR70 pKa = 11.84LFVRR74 pKa = 11.84RR75 pKa = 11.84LEE77 pKa = 4.15VRR79 pKa = 3.29
Molecular weight: 9.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1582622 |
33 |
4135 |
315.5 |
34.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.237 ± 0.044 | 0.893 ± 0.01 |
5.952 ± 0.032 | 6.101 ± 0.039 |
3.996 ± 0.025 | 8.495 ± 0.039 |
1.952 ± 0.018 | 5.452 ± 0.023 |
3.665 ± 0.026 | 10.213 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.523 ± 0.016 | 2.886 ± 0.02 |
4.844 ± 0.028 | 3.181 ± 0.021 |
6.345 ± 0.038 | 5.844 ± 0.027 |
5.477 ± 0.028 | 7.369 ± 0.024 |
1.276 ± 0.013 | 2.299 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
