
Cycloclasticus sp. symbiont of Bathymodiolus heckerae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Piscirickettsiaceae; Cycloclasticus; unclassified Cycloclasticus
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
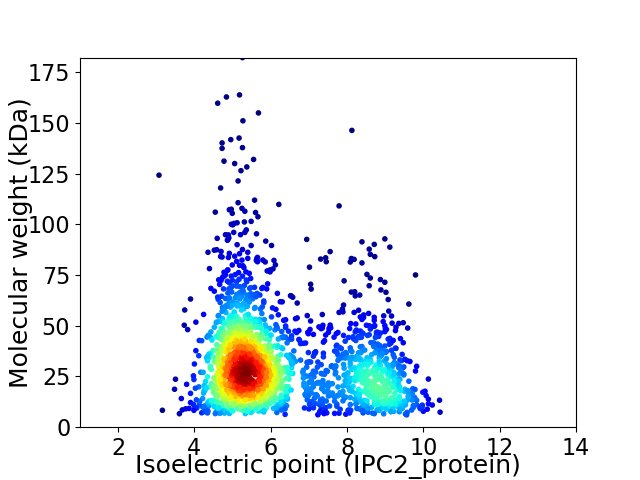
Virtual 2D-PAGE plot for 2010 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X1QTT5|A0A1X1QTT5_9GAMM 50S ribosomal protein L11 OS=Cycloclasticus sp. symbiont of Bathymodiolus heckerae OX=1284675 GN=rplK PE=3 SV=1
MM1 pKa = 7.44RR2 pKa = 11.84TFNKK6 pKa = 9.97KK7 pKa = 9.54PLALGVAAGMAMLVASTGAMAISAGTDD34 pKa = 3.25LNLSGKK40 pKa = 9.8ARR42 pKa = 11.84AGGMAGAAYY51 pKa = 8.83TMPQEE56 pKa = 3.77ASAAVFGNPATLSQFKK72 pKa = 9.99GINMNFGASWLSLSGVEE89 pKa = 4.84NVQTDD94 pKa = 3.66SLGGSNTSTSDD105 pKa = 2.9ADD107 pKa = 3.56NYY109 pKa = 10.25IIPDD113 pKa = 4.22FGLTIQVSPNLVIGTGLEE131 pKa = 3.73VDD133 pKa = 3.84AGLGADD139 pKa = 4.35YY140 pKa = 10.85RR141 pKa = 11.84DD142 pKa = 4.21DD143 pKa = 5.63PINLLPGAGDD153 pKa = 3.38FTLPLLVEE161 pKa = 4.61VISFNANLAAAYY173 pKa = 8.67QATDD177 pKa = 3.53QLSLGASVTVGFGLAQLGTAGATQGLTALNGLVNTGGNGAGAGIAVGPDD226 pKa = 2.63ILADD230 pKa = 4.23FGGTTSSVHH239 pKa = 7.17DD240 pKa = 3.64IGFGASLGAVYY251 pKa = 9.69EE252 pKa = 4.25VQEE255 pKa = 4.35GVLVSATVKK264 pKa = 10.98SPVEE268 pKa = 3.9YY269 pKa = 10.2NFKK272 pKa = 10.9NIIHH276 pKa = 7.35ADD278 pKa = 3.55PTAIAAAGGSIAADD292 pKa = 4.97GYY294 pKa = 11.63QDD296 pKa = 4.49LTLQQPAEE304 pKa = 4.43VILGIALNDD313 pKa = 3.55VIMPGLLIEE322 pKa = 5.95ADD324 pKa = 4.53AIWKK328 pKa = 8.91NWSDD332 pKa = 2.92AHH334 pKa = 7.77AYY336 pKa = 8.84EE337 pKa = 5.58DD338 pKa = 4.21AYY340 pKa = 10.57EE341 pKa = 4.29DD342 pKa = 3.37QWLLAIGAQYY352 pKa = 10.52EE353 pKa = 4.41AGDD356 pKa = 3.34FTYY359 pKa = 10.4RR360 pKa = 11.84VGYY363 pKa = 10.23SYY365 pKa = 11.75AEE367 pKa = 5.49DD368 pKa = 3.62ILSDD372 pKa = 3.87APNGTLSGLDD382 pKa = 3.49GLGSVPLYY390 pKa = 10.41DD391 pKa = 5.12DD392 pKa = 4.7PGANGALGALSVDD405 pKa = 4.06VITIVQNSLLPVIWNHH421 pKa = 6.72TITAGIGYY429 pKa = 9.31KK430 pKa = 8.9ITDD433 pKa = 3.78AVSVDD438 pKa = 3.49AYY440 pKa = 10.46AAYY443 pKa = 10.44AFGEE447 pKa = 4.28NEE449 pKa = 4.38KK450 pKa = 9.79DD451 pKa = 3.53TLSTLSTVADD461 pKa = 3.76TALGGSGLGGTLIEE475 pKa = 4.51NEE477 pKa = 3.87AKK479 pKa = 9.73IDD481 pKa = 3.71YY482 pKa = 10.2EE483 pKa = 4.32LMIGAGINIALPP495 pKa = 3.76
MM1 pKa = 7.44RR2 pKa = 11.84TFNKK6 pKa = 9.97KK7 pKa = 9.54PLALGVAAGMAMLVASTGAMAISAGTDD34 pKa = 3.25LNLSGKK40 pKa = 9.8ARR42 pKa = 11.84AGGMAGAAYY51 pKa = 8.83TMPQEE56 pKa = 3.77ASAAVFGNPATLSQFKK72 pKa = 9.99GINMNFGASWLSLSGVEE89 pKa = 4.84NVQTDD94 pKa = 3.66SLGGSNTSTSDD105 pKa = 2.9ADD107 pKa = 3.56NYY109 pKa = 10.25IIPDD113 pKa = 4.22FGLTIQVSPNLVIGTGLEE131 pKa = 3.73VDD133 pKa = 3.84AGLGADD139 pKa = 4.35YY140 pKa = 10.85RR141 pKa = 11.84DD142 pKa = 4.21DD143 pKa = 5.63PINLLPGAGDD153 pKa = 3.38FTLPLLVEE161 pKa = 4.61VISFNANLAAAYY173 pKa = 8.67QATDD177 pKa = 3.53QLSLGASVTVGFGLAQLGTAGATQGLTALNGLVNTGGNGAGAGIAVGPDD226 pKa = 2.63ILADD230 pKa = 4.23FGGTTSSVHH239 pKa = 7.17DD240 pKa = 3.64IGFGASLGAVYY251 pKa = 9.69EE252 pKa = 4.25VQEE255 pKa = 4.35GVLVSATVKK264 pKa = 10.98SPVEE268 pKa = 3.9YY269 pKa = 10.2NFKK272 pKa = 10.9NIIHH276 pKa = 7.35ADD278 pKa = 3.55PTAIAAAGGSIAADD292 pKa = 4.97GYY294 pKa = 11.63QDD296 pKa = 4.49LTLQQPAEE304 pKa = 4.43VILGIALNDD313 pKa = 3.55VIMPGLLIEE322 pKa = 5.95ADD324 pKa = 4.53AIWKK328 pKa = 8.91NWSDD332 pKa = 2.92AHH334 pKa = 7.77AYY336 pKa = 8.84EE337 pKa = 5.58DD338 pKa = 4.21AYY340 pKa = 10.57EE341 pKa = 4.29DD342 pKa = 3.37QWLLAIGAQYY352 pKa = 10.52EE353 pKa = 4.41AGDD356 pKa = 3.34FTYY359 pKa = 10.4RR360 pKa = 11.84VGYY363 pKa = 10.23SYY365 pKa = 11.75AEE367 pKa = 5.49DD368 pKa = 3.62ILSDD372 pKa = 3.87APNGTLSGLDD382 pKa = 3.49GLGSVPLYY390 pKa = 10.41DD391 pKa = 5.12DD392 pKa = 4.7PGANGALGALSVDD405 pKa = 4.06VITIVQNSLLPVIWNHH421 pKa = 6.72TITAGIGYY429 pKa = 9.31KK430 pKa = 8.9ITDD433 pKa = 3.78AVSVDD438 pKa = 3.49AYY440 pKa = 10.46AAYY443 pKa = 10.44AFGEE447 pKa = 4.28NEE449 pKa = 4.38KK450 pKa = 9.79DD451 pKa = 3.53TLSTLSTVADD461 pKa = 3.76TALGGSGLGGTLIEE475 pKa = 4.51NEE477 pKa = 3.87AKK479 pKa = 9.73IDD481 pKa = 3.71YY482 pKa = 10.2EE483 pKa = 4.32LMIGAGINIALPP495 pKa = 3.76
Molecular weight: 50.25 kDa
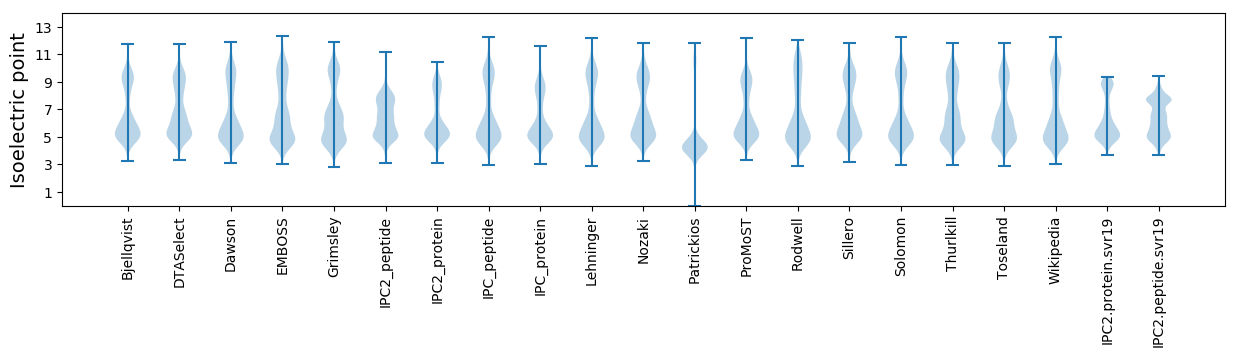
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X1QUG2|A0A1X1QUG2_9GAMM Transposase OS=Cycloclasticus sp. symbiont of Bathymodiolus heckerae OX=1284675 GN=A6F70_09765 PE=4 SV=1
MM1 pKa = 7.61LGLGCPKK8 pKa = 10.21QAFAKK13 pKa = 10.05IATPRR18 pKa = 11.84GTRR21 pKa = 11.84GIRR24 pKa = 11.84LGQQLSLRR32 pKa = 11.84QVNRR36 pKa = 11.84VFVSLRR42 pKa = 11.84DD43 pKa = 3.4KK44 pKa = 11.11LGWVNRR50 pKa = 11.84GAHH53 pKa = 6.18DD54 pKa = 3.74APRR57 pKa = 11.84IHH59 pKa = 7.52DD60 pKa = 3.78LRR62 pKa = 11.84HH63 pKa = 4.95TFVVRR68 pKa = 11.84RR69 pKa = 11.84VMLWHH74 pKa = 5.82TQGINIDD81 pKa = 3.73QQMLALSTYY90 pKa = 9.46VGHH93 pKa = 7.57AMITNTYY100 pKa = 8.62WYY102 pKa = 8.6LTGVPQLMAVAAEE115 pKa = 4.1KK116 pKa = 10.89FEE118 pKa = 5.01AFTQIQEE125 pKa = 4.14ASHH128 pKa = 5.29VV129 pKa = 3.7
MM1 pKa = 7.61LGLGCPKK8 pKa = 10.21QAFAKK13 pKa = 10.05IATPRR18 pKa = 11.84GTRR21 pKa = 11.84GIRR24 pKa = 11.84LGQQLSLRR32 pKa = 11.84QVNRR36 pKa = 11.84VFVSLRR42 pKa = 11.84DD43 pKa = 3.4KK44 pKa = 11.11LGWVNRR50 pKa = 11.84GAHH53 pKa = 6.18DD54 pKa = 3.74APRR57 pKa = 11.84IHH59 pKa = 7.52DD60 pKa = 3.78LRR62 pKa = 11.84HH63 pKa = 4.95TFVVRR68 pKa = 11.84RR69 pKa = 11.84VMLWHH74 pKa = 5.82TQGINIDD81 pKa = 3.73QQMLALSTYY90 pKa = 9.46VGHH93 pKa = 7.57AMITNTYY100 pKa = 8.62WYY102 pKa = 8.6LTGVPQLMAVAAEE115 pKa = 4.1KK116 pKa = 10.89FEE118 pKa = 5.01AFTQIQEE125 pKa = 4.14ASHH128 pKa = 5.29VV129 pKa = 3.7
Molecular weight: 14.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
614684 |
51 |
1614 |
305.8 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.393 ± 0.056 | 1.081 ± 0.02 |
5.707 ± 0.043 | 6.36 ± 0.061 |
4.049 ± 0.039 | 6.956 ± 0.061 |
2.271 ± 0.026 | 6.775 ± 0.04 |
6.041 ± 0.051 | 10.329 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.571 ± 0.032 | 4.286 ± 0.037 |
3.831 ± 0.035 | 4.021 ± 0.043 |
4.501 ± 0.043 | 6.671 ± 0.038 |
5.301 ± 0.038 | 6.916 ± 0.047 |
1.127 ± 0.021 | 2.812 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
