
Rhinocladiella mackenziei CBS 650.93
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Rhinocladiella; Rhinocladiella mackenziei
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
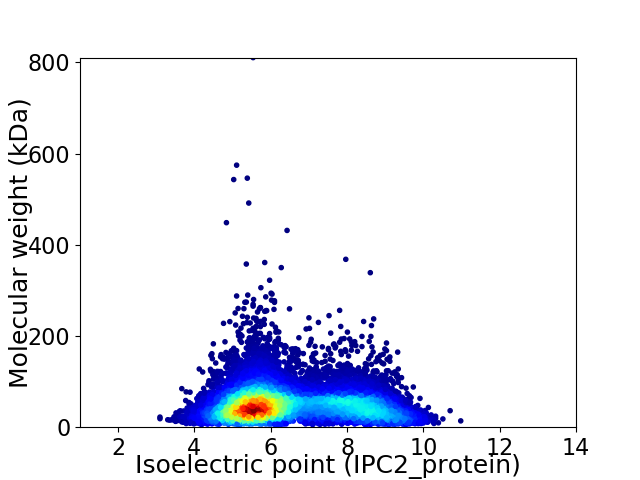
Virtual 2D-PAGE plot for 11378 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0D2IUW8|A0A0D2IUW8_9EURO Rhinocladiella mackenziei CBS 650.93 unplaced genomic scaffold supercont1.1 whole genome shotgun sequence OS=Rhinocladiella mackenziei CBS 650.93 OX=1442369 GN=Z518_00965 PE=4 SV=1
MM1 pKa = 7.75HH2 pKa = 7.39SFTVLLAVSAFASHH16 pKa = 6.72SLAKK20 pKa = 9.83PIPASDD26 pKa = 3.22VSTKK30 pKa = 10.59KK31 pKa = 10.88GFTVKK36 pKa = 10.25QAIAKK41 pKa = 7.52PFQAGPVVLQKK52 pKa = 9.76TYY54 pKa = 11.11NKK56 pKa = 8.92YY57 pKa = 6.29TTAVPADD64 pKa = 3.91VKK66 pKa = 10.93AAAADD71 pKa = 4.04GSVTATPEE79 pKa = 3.97QYY81 pKa = 10.38DD82 pKa = 3.78AEE84 pKa = 4.38YY85 pKa = 10.32LSPVSIGGQTLNLDD99 pKa = 4.16FDD101 pKa = 4.24TGSADD106 pKa = 2.96LWVFSSEE113 pKa = 4.48LPSSQQSGHH122 pKa = 6.24SIYY125 pKa = 10.89DD126 pKa = 3.41PSKK129 pKa = 10.68SDD131 pKa = 3.2TAKK134 pKa = 10.35EE135 pKa = 3.96LEE137 pKa = 4.22GATWNISYY145 pKa = 11.03GDD147 pKa = 3.67GSGASGNVYY156 pKa = 10.06TDD158 pKa = 3.39TVDD161 pKa = 3.18VGGTTVTGQAVEE173 pKa = 4.31LAAQISAQFQQDD185 pKa = 3.53EE186 pKa = 4.68NNDD189 pKa = 3.65GLLGLAFSSINTVQPDD205 pKa = 3.47QQTTFFDD212 pKa = 3.56TAIQQGVLSQNLFTVDD228 pKa = 3.9LKK230 pKa = 11.06KK231 pKa = 11.08GEE233 pKa = 4.41PGTYY237 pKa = 10.33DD238 pKa = 3.37FGFIDD243 pKa = 4.49DD244 pKa = 4.63SKK246 pKa = 9.85HH247 pKa = 5.86TGDD250 pKa = 3.05ITYY253 pKa = 9.54TPVDD257 pKa = 3.53TANGFWEE264 pKa = 4.71FTGTGYY270 pKa = 11.33AVGDD274 pKa = 3.91GSFQQQSIDD283 pKa = 3.95AIADD287 pKa = 3.46TGTTLLLMDD296 pKa = 5.35DD297 pKa = 5.07SIVSAYY303 pKa = 10.12YY304 pKa = 10.47DD305 pKa = 3.31QVDD308 pKa = 3.61GAKK311 pKa = 10.19YY312 pKa = 10.87DD313 pKa = 3.85NTQGGYY319 pKa = 7.62TFPCSADD326 pKa = 3.45LPDD329 pKa = 3.85FAVGIEE335 pKa = 4.78DD336 pKa = 3.53YY337 pKa = 10.85HH338 pKa = 8.3AVIPGSFMNFAPADD352 pKa = 3.49SSTCYY357 pKa = 10.5GGLQSNEE364 pKa = 4.07GIGFSIYY371 pKa = 11.02GDD373 pKa = 3.29IFLKK377 pKa = 10.87AVFAVFDD384 pKa = 4.03SDD386 pKa = 3.46NTQLGFAPKK395 pKa = 10.41DD396 pKa = 3.43LL397 pKa = 4.54
MM1 pKa = 7.75HH2 pKa = 7.39SFTVLLAVSAFASHH16 pKa = 6.72SLAKK20 pKa = 9.83PIPASDD26 pKa = 3.22VSTKK30 pKa = 10.59KK31 pKa = 10.88GFTVKK36 pKa = 10.25QAIAKK41 pKa = 7.52PFQAGPVVLQKK52 pKa = 9.76TYY54 pKa = 11.11NKK56 pKa = 8.92YY57 pKa = 6.29TTAVPADD64 pKa = 3.91VKK66 pKa = 10.93AAAADD71 pKa = 4.04GSVTATPEE79 pKa = 3.97QYY81 pKa = 10.38DD82 pKa = 3.78AEE84 pKa = 4.38YY85 pKa = 10.32LSPVSIGGQTLNLDD99 pKa = 4.16FDD101 pKa = 4.24TGSADD106 pKa = 2.96LWVFSSEE113 pKa = 4.48LPSSQQSGHH122 pKa = 6.24SIYY125 pKa = 10.89DD126 pKa = 3.41PSKK129 pKa = 10.68SDD131 pKa = 3.2TAKK134 pKa = 10.35EE135 pKa = 3.96LEE137 pKa = 4.22GATWNISYY145 pKa = 11.03GDD147 pKa = 3.67GSGASGNVYY156 pKa = 10.06TDD158 pKa = 3.39TVDD161 pKa = 3.18VGGTTVTGQAVEE173 pKa = 4.31LAAQISAQFQQDD185 pKa = 3.53EE186 pKa = 4.68NNDD189 pKa = 3.65GLLGLAFSSINTVQPDD205 pKa = 3.47QQTTFFDD212 pKa = 3.56TAIQQGVLSQNLFTVDD228 pKa = 3.9LKK230 pKa = 11.06KK231 pKa = 11.08GEE233 pKa = 4.41PGTYY237 pKa = 10.33DD238 pKa = 3.37FGFIDD243 pKa = 4.49DD244 pKa = 4.63SKK246 pKa = 9.85HH247 pKa = 5.86TGDD250 pKa = 3.05ITYY253 pKa = 9.54TPVDD257 pKa = 3.53TANGFWEE264 pKa = 4.71FTGTGYY270 pKa = 11.33AVGDD274 pKa = 3.91GSFQQQSIDD283 pKa = 3.95AIADD287 pKa = 3.46TGTTLLLMDD296 pKa = 5.35DD297 pKa = 5.07SIVSAYY303 pKa = 10.12YY304 pKa = 10.47DD305 pKa = 3.31QVDD308 pKa = 3.61GAKK311 pKa = 10.19YY312 pKa = 10.87DD313 pKa = 3.85NTQGGYY319 pKa = 7.62TFPCSADD326 pKa = 3.45LPDD329 pKa = 3.85FAVGIEE335 pKa = 4.78DD336 pKa = 3.53YY337 pKa = 10.85HH338 pKa = 8.3AVIPGSFMNFAPADD352 pKa = 3.49SSTCYY357 pKa = 10.5GGLQSNEE364 pKa = 4.07GIGFSIYY371 pKa = 11.02GDD373 pKa = 3.29IFLKK377 pKa = 10.87AVFAVFDD384 pKa = 4.03SDD386 pKa = 3.46NTQLGFAPKK395 pKa = 10.41DD396 pKa = 3.43LL397 pKa = 4.54
Molecular weight: 41.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2J7K2|A0A0D2J7K2_9EURO Rhinocladiella mackenziei CBS 650.93 unplaced genomic scaffold supercont1.4 whole genome shotgun sequence OS=Rhinocladiella mackenziei CBS 650.93 OX=1442369 GN=Z518_05877 PE=4 SV=1
MM1 pKa = 7.53ALGRR5 pKa = 11.84RR6 pKa = 11.84TRR8 pKa = 11.84HH9 pKa = 5.74HH10 pKa = 6.8SSGTTVTTTTTTTHH24 pKa = 6.64HH25 pKa = 6.68PPGTKK30 pKa = 9.84VSPSGQVRR38 pKa = 11.84TKK40 pKa = 8.57PTFMQRR46 pKa = 11.84LRR48 pKa = 11.84GNTTPRR54 pKa = 11.84TTTARR59 pKa = 11.84PARR62 pKa = 11.84QRR64 pKa = 11.84RR65 pKa = 11.84WGGTRR70 pKa = 11.84TRR72 pKa = 11.84RR73 pKa = 11.84TPATATTPSRR83 pKa = 11.84KK84 pKa = 8.13TSIGDD89 pKa = 3.66KK90 pKa = 10.84VSGAMLKK97 pKa = 10.78LKK99 pKa = 9.49GTVTRR104 pKa = 11.84RR105 pKa = 11.84PGQKK109 pKa = 9.82AAGTRR114 pKa = 11.84RR115 pKa = 11.84MHH117 pKa = 5.82GTDD120 pKa = 3.0GSGTRR125 pKa = 11.84RR126 pKa = 11.84YY127 pKa = 10.03
MM1 pKa = 7.53ALGRR5 pKa = 11.84RR6 pKa = 11.84TRR8 pKa = 11.84HH9 pKa = 5.74HH10 pKa = 6.8SSGTTVTTTTTTTHH24 pKa = 6.64HH25 pKa = 6.68PPGTKK30 pKa = 9.84VSPSGQVRR38 pKa = 11.84TKK40 pKa = 8.57PTFMQRR46 pKa = 11.84LRR48 pKa = 11.84GNTTPRR54 pKa = 11.84TTTARR59 pKa = 11.84PARR62 pKa = 11.84QRR64 pKa = 11.84RR65 pKa = 11.84WGGTRR70 pKa = 11.84TRR72 pKa = 11.84RR73 pKa = 11.84TPATATTPSRR83 pKa = 11.84KK84 pKa = 8.13TSIGDD89 pKa = 3.66KK90 pKa = 10.84VSGAMLKK97 pKa = 10.78LKK99 pKa = 9.49GTVTRR104 pKa = 11.84RR105 pKa = 11.84PGQKK109 pKa = 9.82AAGTRR114 pKa = 11.84RR115 pKa = 11.84MHH117 pKa = 5.82GTDD120 pKa = 3.0GSGTRR125 pKa = 11.84RR126 pKa = 11.84YY127 pKa = 10.03
Molecular weight: 13.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5555827 |
50 |
7334 |
488.3 |
54.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.257 ± 0.018 | 1.228 ± 0.008 |
5.714 ± 0.015 | 6.164 ± 0.018 |
3.789 ± 0.013 | 6.738 ± 0.02 |
2.446 ± 0.01 | 5.086 ± 0.015 |
4.83 ± 0.018 | 8.968 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.221 ± 0.009 | 3.697 ± 0.01 |
6.045 ± 0.021 | 4.05 ± 0.014 |
6.237 ± 0.017 | 8.2 ± 0.026 |
5.979 ± 0.014 | 6.089 ± 0.014 |
1.484 ± 0.009 | 2.777 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
