
Stackebrandtia albiflava
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Glycomycetales; Glycomycetaceae; Stackebrandtia
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
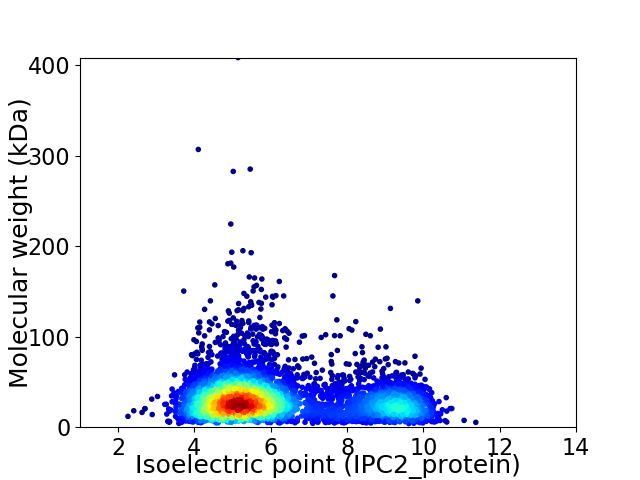
Virtual 2D-PAGE plot for 5212 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A562UQ83|A0A562UQ83_9ACTN Peptide/nickel transport system ATP-binding protein/oligopeptide transport system ATP-binding protein OS=Stackebrandtia albiflava OX=406432 GN=LX16_4916 PE=4 SV=1
MM1 pKa = 6.86TRR3 pKa = 11.84IRR5 pKa = 11.84RR6 pKa = 11.84LSYY9 pKa = 10.26IAMAAALSFTAAACTSQGPSNDD31 pKa = 3.33AQNQTEE37 pKa = 4.33TQDD40 pKa = 3.4VVAGGEE46 pKa = 3.86ITIAATSPITDD57 pKa = 3.76WNPISANGDD66 pKa = 3.33TTGQRR71 pKa = 11.84QQQWPLYY78 pKa = 7.17PHH80 pKa = 7.44PFLTLPDD87 pKa = 3.53TSVVINEE94 pKa = 4.29ALLEE98 pKa = 4.26KK99 pKa = 10.58ADD101 pKa = 3.93VVEE104 pKa = 4.23TDD106 pKa = 3.26PMTVEE111 pKa = 4.59YY112 pKa = 10.66VLQDD116 pKa = 3.27GVQWSDD122 pKa = 3.19GTPIGVDD129 pKa = 2.98DD130 pKa = 4.63FVYY133 pKa = 9.37TQAVQDD139 pKa = 3.92PSKK142 pKa = 10.85CAEE145 pKa = 4.06CLAAFTEE152 pKa = 5.37GYY154 pKa = 10.67SDD156 pKa = 3.18ITSIEE161 pKa = 3.73ASEE164 pKa = 4.42DD165 pKa = 3.22GKK167 pKa = 9.59TITMVYY173 pKa = 10.07SEE175 pKa = 5.32PFSEE179 pKa = 3.8WRR181 pKa = 11.84ALFNYY186 pKa = 9.65ILPAHH191 pKa = 5.61VAEE194 pKa = 5.52GYY196 pKa = 11.16GDD198 pKa = 4.16LATSFNEE205 pKa = 4.25GFSKK209 pKa = 10.63NVPEE213 pKa = 4.55FSGGPYY219 pKa = 9.86IIQDD223 pKa = 3.69YY224 pKa = 10.11TDD226 pKa = 5.2GISMTMVPNPNWYY239 pKa = 10.49GEE241 pKa = 4.25GPNLEE246 pKa = 4.75TITTRR251 pKa = 11.84YY252 pKa = 8.0ITGQGEE258 pKa = 4.29QLTALQSGEE267 pKa = 4.15VQLVYY272 pKa = 9.89ATPTVDD278 pKa = 3.37TMDD281 pKa = 4.14QARR284 pKa = 11.84QLPNMTINTGSTLTYY299 pKa = 9.44YY300 pKa = 10.66HH301 pKa = 7.21LGMKK305 pKa = 7.36TTGDD309 pKa = 3.77VMSDD313 pKa = 3.19PALRR317 pKa = 11.84SAIATALDD325 pKa = 3.88LGDD328 pKa = 3.45MRR330 pKa = 11.84EE331 pKa = 3.84RR332 pKa = 11.84TIGQFAPDD340 pKa = 3.86VPQMKK345 pKa = 9.97SSVYY349 pKa = 10.59VPGQQIGGQEE359 pKa = 3.98AYY361 pKa = 9.96RR362 pKa = 11.84DD363 pKa = 3.58NMTDD367 pKa = 3.35LGIGAGDD374 pKa = 3.45VEE376 pKa = 4.58GAIGILEE383 pKa = 4.27DD384 pKa = 3.53AGYY387 pKa = 8.98TITDD391 pKa = 3.6EE392 pKa = 5.36QLIQPDD398 pKa = 4.19GTPLRR403 pKa = 11.84DD404 pKa = 3.84LSFLTLGTDD413 pKa = 3.62VLRR416 pKa = 11.84MEE418 pKa = 4.84VAQIAQQQLLALGITINIDD437 pKa = 3.14AADD440 pKa = 4.05GARR443 pKa = 11.84YY444 pKa = 9.83SPALRR449 pKa = 11.84EE450 pKa = 3.94GDD452 pKa = 3.92FDD454 pKa = 5.65LMATGTALDD463 pKa = 4.75LGPLSMQQWYY473 pKa = 8.49GTDD476 pKa = 3.32APRR479 pKa = 11.84SFGYY483 pKa = 10.69SNPEE487 pKa = 3.25ADD489 pKa = 4.59ALLEE493 pKa = 4.25AAASEE498 pKa = 4.69LDD500 pKa = 3.37PAAQIEE506 pKa = 4.63LMNEE510 pKa = 4.16LDD512 pKa = 4.57RR513 pKa = 11.84VLLGDD518 pKa = 4.01GVVLPLFGTPMMAVYY533 pKa = 10.17PNTYY537 pKa = 10.52GNIFINPSKK546 pKa = 10.97YY547 pKa = 8.03GTTMNVEE554 pKa = 3.93EE555 pKa = 4.68WGLLAEE561 pKa = 5.34PGTSS565 pKa = 3.06
MM1 pKa = 6.86TRR3 pKa = 11.84IRR5 pKa = 11.84RR6 pKa = 11.84LSYY9 pKa = 10.26IAMAAALSFTAAACTSQGPSNDD31 pKa = 3.33AQNQTEE37 pKa = 4.33TQDD40 pKa = 3.4VVAGGEE46 pKa = 3.86ITIAATSPITDD57 pKa = 3.76WNPISANGDD66 pKa = 3.33TTGQRR71 pKa = 11.84QQQWPLYY78 pKa = 7.17PHH80 pKa = 7.44PFLTLPDD87 pKa = 3.53TSVVINEE94 pKa = 4.29ALLEE98 pKa = 4.26KK99 pKa = 10.58ADD101 pKa = 3.93VVEE104 pKa = 4.23TDD106 pKa = 3.26PMTVEE111 pKa = 4.59YY112 pKa = 10.66VLQDD116 pKa = 3.27GVQWSDD122 pKa = 3.19GTPIGVDD129 pKa = 2.98DD130 pKa = 4.63FVYY133 pKa = 9.37TQAVQDD139 pKa = 3.92PSKK142 pKa = 10.85CAEE145 pKa = 4.06CLAAFTEE152 pKa = 5.37GYY154 pKa = 10.67SDD156 pKa = 3.18ITSIEE161 pKa = 3.73ASEE164 pKa = 4.42DD165 pKa = 3.22GKK167 pKa = 9.59TITMVYY173 pKa = 10.07SEE175 pKa = 5.32PFSEE179 pKa = 3.8WRR181 pKa = 11.84ALFNYY186 pKa = 9.65ILPAHH191 pKa = 5.61VAEE194 pKa = 5.52GYY196 pKa = 11.16GDD198 pKa = 4.16LATSFNEE205 pKa = 4.25GFSKK209 pKa = 10.63NVPEE213 pKa = 4.55FSGGPYY219 pKa = 9.86IIQDD223 pKa = 3.69YY224 pKa = 10.11TDD226 pKa = 5.2GISMTMVPNPNWYY239 pKa = 10.49GEE241 pKa = 4.25GPNLEE246 pKa = 4.75TITTRR251 pKa = 11.84YY252 pKa = 8.0ITGQGEE258 pKa = 4.29QLTALQSGEE267 pKa = 4.15VQLVYY272 pKa = 9.89ATPTVDD278 pKa = 3.37TMDD281 pKa = 4.14QARR284 pKa = 11.84QLPNMTINTGSTLTYY299 pKa = 9.44YY300 pKa = 10.66HH301 pKa = 7.21LGMKK305 pKa = 7.36TTGDD309 pKa = 3.77VMSDD313 pKa = 3.19PALRR317 pKa = 11.84SAIATALDD325 pKa = 3.88LGDD328 pKa = 3.45MRR330 pKa = 11.84EE331 pKa = 3.84RR332 pKa = 11.84TIGQFAPDD340 pKa = 3.86VPQMKK345 pKa = 9.97SSVYY349 pKa = 10.59VPGQQIGGQEE359 pKa = 3.98AYY361 pKa = 9.96RR362 pKa = 11.84DD363 pKa = 3.58NMTDD367 pKa = 3.35LGIGAGDD374 pKa = 3.45VEE376 pKa = 4.58GAIGILEE383 pKa = 4.27DD384 pKa = 3.53AGYY387 pKa = 8.98TITDD391 pKa = 3.6EE392 pKa = 5.36QLIQPDD398 pKa = 4.19GTPLRR403 pKa = 11.84DD404 pKa = 3.84LSFLTLGTDD413 pKa = 3.62VLRR416 pKa = 11.84MEE418 pKa = 4.84VAQIAQQQLLALGITINIDD437 pKa = 3.14AADD440 pKa = 4.05GARR443 pKa = 11.84YY444 pKa = 9.83SPALRR449 pKa = 11.84EE450 pKa = 3.94GDD452 pKa = 3.92FDD454 pKa = 5.65LMATGTALDD463 pKa = 4.75LGPLSMQQWYY473 pKa = 8.49GTDD476 pKa = 3.32APRR479 pKa = 11.84SFGYY483 pKa = 10.69SNPEE487 pKa = 3.25ADD489 pKa = 4.59ALLEE493 pKa = 4.25AAASEE498 pKa = 4.69LDD500 pKa = 3.37PAAQIEE506 pKa = 4.63LMNEE510 pKa = 4.16LDD512 pKa = 4.57RR513 pKa = 11.84VLLGDD518 pKa = 4.01GVVLPLFGTPMMAVYY533 pKa = 10.17PNTYY537 pKa = 10.52GNIFINPSKK546 pKa = 10.97YY547 pKa = 8.03GTTMNVEE554 pKa = 3.93EE555 pKa = 4.68WGLLAEE561 pKa = 5.34PGTSS565 pKa = 3.06
Molecular weight: 60.94 kDa
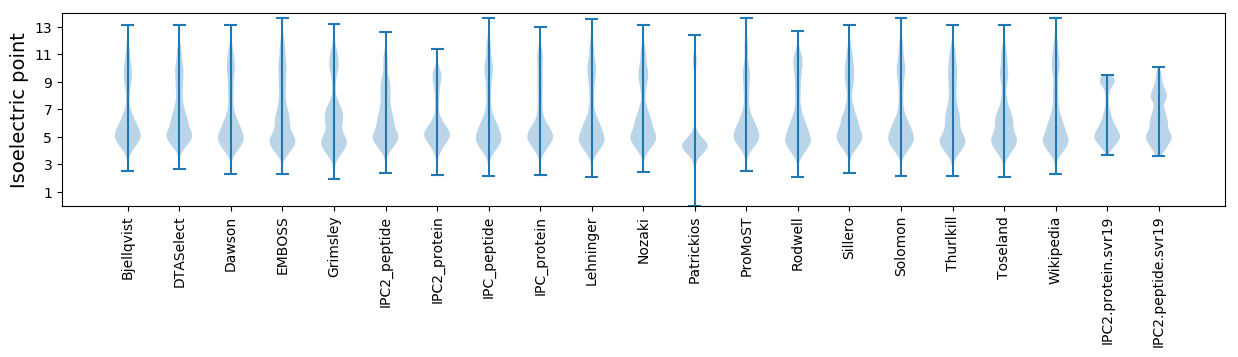
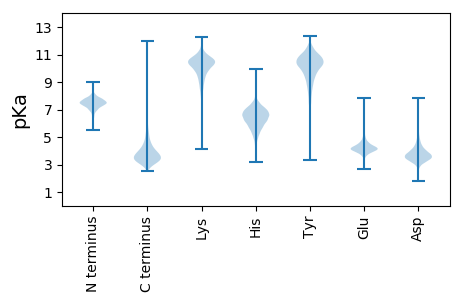
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A562V129|A0A562V129_9ACTN DNA replication and repair protein RecF OS=Stackebrandtia albiflava OX=406432 GN=recF PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LTAA45 pKa = 4.29
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LTAA45 pKa = 4.29
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1617362 |
37 |
3801 |
310.3 |
33.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.179 ± 0.049 | 0.807 ± 0.01 |
6.61 ± 0.034 | 5.673 ± 0.033 |
2.826 ± 0.018 | 9.155 ± 0.035 |
2.324 ± 0.019 | 3.456 ± 0.021 |
1.6 ± 0.02 | 9.98 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.965 ± 0.015 | 1.717 ± 0.015 |
6.047 ± 0.035 | 2.348 ± 0.02 |
8.336 ± 0.042 | 4.928 ± 0.022 |
6.369 ± 0.031 | 9.011 ± 0.036 |
1.612 ± 0.014 | 2.059 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
