
Macrophomina phaseolina tobamo-like virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; unclassified Virgaviridae
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
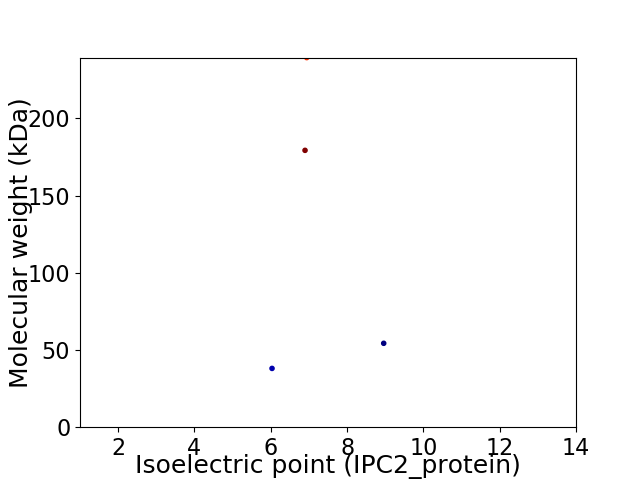
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A088CPQ6|A0A088CPQ6_9VIRU Methyltransferase/RNA helicase OS=Macrophomina phaseolina tobamo-like virus OX=1527523 PE=4 SV=1
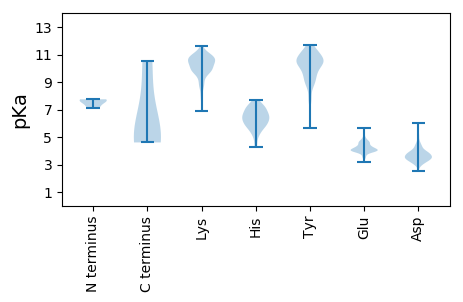
MM1 pKa = 7.12SQQIKK6 pKa = 10.8CPVCQKK12 pKa = 11.1DD13 pKa = 3.69FDD15 pKa = 4.33RR16 pKa = 11.84KK17 pKa = 9.27QAEE20 pKa = 3.94LHH22 pKa = 5.81IAEE25 pKa = 4.8AHH27 pKa = 5.88SSPDD31 pKa = 3.23KK32 pKa = 11.22KK33 pKa = 10.44EE34 pKa = 4.05WPDD37 pKa = 3.65LPDD40 pKa = 4.14GNTLTEE46 pKa = 4.03AEE48 pKa = 4.98FIRR51 pKa = 11.84MALEE55 pKa = 4.22KK56 pKa = 9.93PQEE59 pKa = 3.91IGVAIDD65 pKa = 3.37QPVTVVPPSTIFEE78 pKa = 5.11DD79 pKa = 3.14IGIKK83 pKa = 10.16VDD85 pKa = 4.07DD86 pKa = 4.42FQHH89 pKa = 7.53VDD91 pKa = 2.75ITTMAGATLTSSEE104 pKa = 3.98AKK106 pKa = 9.74MLFNVLEE113 pKa = 3.85EE114 pKa = 4.11HH115 pKa = 6.92LAFKK119 pKa = 10.48INRR122 pKa = 11.84AAFVANFIDD131 pKa = 3.31WGIRR135 pKa = 11.84TSFTEE140 pKa = 4.0EE141 pKa = 3.9LEE143 pKa = 4.31EE144 pKa = 5.23AGGFSVASNPVSGKK158 pKa = 8.99LQEE161 pKa = 4.68RR162 pKa = 11.84YY163 pKa = 6.71MTVLEE168 pKa = 3.84MHH170 pKa = 7.06KK171 pKa = 10.41IITEE175 pKa = 3.85YY176 pKa = 10.36FAAKK180 pKa = 8.58GTGRR184 pKa = 11.84SFTVRR189 pKa = 11.84RR190 pKa = 11.84FGRR193 pKa = 11.84FLAPKK198 pKa = 9.96IPDD201 pKa = 2.99ICDD204 pKa = 3.14QVEE207 pKa = 4.21KK208 pKa = 11.11LKK210 pKa = 10.34MYY212 pKa = 7.6YY213 pKa = 7.71TTGSPMSRR221 pKa = 11.84RR222 pKa = 11.84LGVRR226 pKa = 11.84NNLFLTVTSIYY237 pKa = 10.71EE238 pKa = 4.01YY239 pKa = 10.46IKK241 pKa = 10.55PFNKK245 pKa = 7.59WTRR248 pKa = 11.84DD249 pKa = 3.13EE250 pKa = 4.06KK251 pKa = 11.31AAWEE255 pKa = 4.04ALNRR259 pKa = 11.84TVTKK263 pKa = 10.3QPRR266 pKa = 11.84AQDD269 pKa = 3.49TSFVPQDD276 pKa = 3.58LRR278 pKa = 11.84PRR280 pKa = 11.84PYY282 pKa = 9.8EE283 pKa = 4.01AEE285 pKa = 4.08TLVEE289 pKa = 4.13TRR291 pKa = 11.84STDD294 pKa = 3.4FARR297 pKa = 11.84RR298 pKa = 11.84HH299 pKa = 5.64DD300 pKa = 4.31TDD302 pKa = 3.39PLKK305 pKa = 10.98AVFGQPAAVQGGYY318 pKa = 10.85GKK320 pKa = 10.57GAEE323 pKa = 4.3IFEE326 pKa = 4.31KK327 pKa = 10.25MRR329 pKa = 11.84TGSSGGKK336 pKa = 8.81NAA338 pKa = 4.61
MM1 pKa = 7.12SQQIKK6 pKa = 10.8CPVCQKK12 pKa = 11.1DD13 pKa = 3.69FDD15 pKa = 4.33RR16 pKa = 11.84KK17 pKa = 9.27QAEE20 pKa = 3.94LHH22 pKa = 5.81IAEE25 pKa = 4.8AHH27 pKa = 5.88SSPDD31 pKa = 3.23KK32 pKa = 11.22KK33 pKa = 10.44EE34 pKa = 4.05WPDD37 pKa = 3.65LPDD40 pKa = 4.14GNTLTEE46 pKa = 4.03AEE48 pKa = 4.98FIRR51 pKa = 11.84MALEE55 pKa = 4.22KK56 pKa = 9.93PQEE59 pKa = 3.91IGVAIDD65 pKa = 3.37QPVTVVPPSTIFEE78 pKa = 5.11DD79 pKa = 3.14IGIKK83 pKa = 10.16VDD85 pKa = 4.07DD86 pKa = 4.42FQHH89 pKa = 7.53VDD91 pKa = 2.75ITTMAGATLTSSEE104 pKa = 3.98AKK106 pKa = 9.74MLFNVLEE113 pKa = 3.85EE114 pKa = 4.11HH115 pKa = 6.92LAFKK119 pKa = 10.48INRR122 pKa = 11.84AAFVANFIDD131 pKa = 3.31WGIRR135 pKa = 11.84TSFTEE140 pKa = 4.0EE141 pKa = 3.9LEE143 pKa = 4.31EE144 pKa = 5.23AGGFSVASNPVSGKK158 pKa = 8.99LQEE161 pKa = 4.68RR162 pKa = 11.84YY163 pKa = 6.71MTVLEE168 pKa = 3.84MHH170 pKa = 7.06KK171 pKa = 10.41IITEE175 pKa = 3.85YY176 pKa = 10.36FAAKK180 pKa = 8.58GTGRR184 pKa = 11.84SFTVRR189 pKa = 11.84RR190 pKa = 11.84FGRR193 pKa = 11.84FLAPKK198 pKa = 9.96IPDD201 pKa = 2.99ICDD204 pKa = 3.14QVEE207 pKa = 4.21KK208 pKa = 11.11LKK210 pKa = 10.34MYY212 pKa = 7.6YY213 pKa = 7.71TTGSPMSRR221 pKa = 11.84RR222 pKa = 11.84LGVRR226 pKa = 11.84NNLFLTVTSIYY237 pKa = 10.71EE238 pKa = 4.01YY239 pKa = 10.46IKK241 pKa = 10.55PFNKK245 pKa = 7.59WTRR248 pKa = 11.84DD249 pKa = 3.13EE250 pKa = 4.06KK251 pKa = 11.31AAWEE255 pKa = 4.04ALNRR259 pKa = 11.84TVTKK263 pKa = 10.3QPRR266 pKa = 11.84AQDD269 pKa = 3.49TSFVPQDD276 pKa = 3.58LRR278 pKa = 11.84PRR280 pKa = 11.84PYY282 pKa = 9.8EE283 pKa = 4.01AEE285 pKa = 4.08TLVEE289 pKa = 4.13TRR291 pKa = 11.84STDD294 pKa = 3.4FARR297 pKa = 11.84RR298 pKa = 11.84HH299 pKa = 5.64DD300 pKa = 4.31TDD302 pKa = 3.39PLKK305 pKa = 10.98AVFGQPAAVQGGYY318 pKa = 10.85GKK320 pKa = 10.57GAEE323 pKa = 4.3IFEE326 pKa = 4.31KK327 pKa = 10.25MRR329 pKa = 11.84TGSSGGKK336 pKa = 8.81NAA338 pKa = 4.61
Molecular weight: 38.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A088CPZ9|A0A088CPZ9_9VIRU RNA helicase OS=Macrophomina phaseolina tobamo-like virus OX=1527523 PE=4 SV=1
MM1 pKa = 7.51AFKK4 pKa = 10.67YY5 pKa = 10.57RR6 pKa = 11.84NNPKK10 pKa = 10.12SFSLPRR16 pKa = 11.84EE17 pKa = 4.2FVAHH21 pKa = 6.22FRR23 pKa = 11.84EE24 pKa = 4.49SQNVDD29 pKa = 2.5TDD31 pKa = 3.37RR32 pKa = 11.84TLFGFQVLFTKK43 pKa = 9.46TKK45 pKa = 9.36KK46 pKa = 10.64KK47 pKa = 8.91EE48 pKa = 3.88WLEE51 pKa = 4.2GGHH54 pKa = 6.81APVTFLRR61 pKa = 11.84FFRR64 pKa = 11.84EE65 pKa = 3.87FGQWVKK71 pKa = 10.9GFSAKK76 pKa = 9.76EE77 pKa = 3.46RR78 pKa = 11.84GAAAIAATKK87 pKa = 10.11HH88 pKa = 6.03KK89 pKa = 10.85GVGLYY94 pKa = 9.42PRR96 pKa = 11.84RR97 pKa = 11.84GVYY100 pKa = 9.78RR101 pKa = 11.84DD102 pKa = 3.15NGDD105 pKa = 3.41YY106 pKa = 10.94LGTYY110 pKa = 8.92QEE112 pKa = 4.54VVSEE116 pKa = 4.15GKK118 pKa = 9.7IDD120 pKa = 3.72PTLLAMGMIDD130 pKa = 3.71VLDD133 pKa = 3.89SSVIEE138 pKa = 4.37SSTSQTSQATSSTTTSKK155 pKa = 10.75KK156 pKa = 10.05ASKK159 pKa = 10.21KK160 pKa = 10.45KK161 pKa = 10.82SFTANSAKK169 pKa = 9.87TSSARR174 pKa = 11.84VTHH177 pKa = 6.49AQAKK181 pKa = 7.83DD182 pKa = 3.53TTQKK186 pKa = 11.14AQVLDD191 pKa = 3.44TSIQKK196 pKa = 8.32RR197 pKa = 11.84TSYY200 pKa = 11.03SQFNLGDD207 pKa = 3.51RR208 pKa = 11.84QRR210 pKa = 11.84PLSFTEE216 pKa = 3.48VHH218 pKa = 5.94VPVARR223 pKa = 11.84GNYY226 pKa = 9.5RR227 pKa = 11.84GVTQKK232 pKa = 10.13IVAGPPGGYY241 pKa = 10.15VITGPTGCGKK251 pKa = 8.87STVALLPLFSGKK263 pKa = 10.37SSVLIVEE270 pKa = 4.64PTQANAANIFHH281 pKa = 6.87EE282 pKa = 4.91FSNVLPTLHH291 pKa = 6.66EE292 pKa = 4.68AGVIPWNVPPVEE304 pKa = 4.36FTAPTTRR311 pKa = 11.84EE312 pKa = 3.68GSYY315 pKa = 11.31GPLSVTTTDD324 pKa = 3.44KK325 pKa = 11.02LLEE328 pKa = 4.03YY329 pKa = 10.35FEE331 pKa = 5.0WKK333 pKa = 10.51GSLPKK338 pKa = 9.97VDD340 pKa = 4.17YY341 pKa = 11.21LIIDD345 pKa = 4.2EE346 pKa = 4.48FHH348 pKa = 6.92LPIPSMVQTVEE359 pKa = 3.96LLRR362 pKa = 11.84TFALVPKK369 pKa = 9.42YY370 pKa = 10.51IFVSATAVGYY380 pKa = 9.8SVNPEE385 pKa = 3.75LPKK388 pKa = 10.75AVTQTWGQLPIGQIPSKK405 pKa = 10.61MEE407 pKa = 4.12GSDD410 pKa = 3.47LDD412 pKa = 3.53PRR414 pKa = 11.84RR415 pKa = 11.84WWKK418 pKa = 10.84RR419 pKa = 11.84GDD421 pKa = 3.43GNVAVVAPSVIVAKK435 pKa = 10.55RR436 pKa = 11.84LFNVYY441 pKa = 10.07RR442 pKa = 11.84DD443 pKa = 3.42WQIRR447 pKa = 11.84AFLITRR453 pKa = 11.84EE454 pKa = 4.39TFVSEE459 pKa = 3.95YY460 pKa = 9.96MKK462 pKa = 10.83AATNYY467 pKa = 10.21RR468 pKa = 11.84SMTTFVLEE476 pKa = 4.39PGVEE480 pKa = 4.1AGVTLCIDD488 pKa = 3.48VLII491 pKa = 4.86
MM1 pKa = 7.51AFKK4 pKa = 10.67YY5 pKa = 10.57RR6 pKa = 11.84NNPKK10 pKa = 10.12SFSLPRR16 pKa = 11.84EE17 pKa = 4.2FVAHH21 pKa = 6.22FRR23 pKa = 11.84EE24 pKa = 4.49SQNVDD29 pKa = 2.5TDD31 pKa = 3.37RR32 pKa = 11.84TLFGFQVLFTKK43 pKa = 9.46TKK45 pKa = 9.36KK46 pKa = 10.64KK47 pKa = 8.91EE48 pKa = 3.88WLEE51 pKa = 4.2GGHH54 pKa = 6.81APVTFLRR61 pKa = 11.84FFRR64 pKa = 11.84EE65 pKa = 3.87FGQWVKK71 pKa = 10.9GFSAKK76 pKa = 9.76EE77 pKa = 3.46RR78 pKa = 11.84GAAAIAATKK87 pKa = 10.11HH88 pKa = 6.03KK89 pKa = 10.85GVGLYY94 pKa = 9.42PRR96 pKa = 11.84RR97 pKa = 11.84GVYY100 pKa = 9.78RR101 pKa = 11.84DD102 pKa = 3.15NGDD105 pKa = 3.41YY106 pKa = 10.94LGTYY110 pKa = 8.92QEE112 pKa = 4.54VVSEE116 pKa = 4.15GKK118 pKa = 9.7IDD120 pKa = 3.72PTLLAMGMIDD130 pKa = 3.71VLDD133 pKa = 3.89SSVIEE138 pKa = 4.37SSTSQTSQATSSTTTSKK155 pKa = 10.75KK156 pKa = 10.05ASKK159 pKa = 10.21KK160 pKa = 10.45KK161 pKa = 10.82SFTANSAKK169 pKa = 9.87TSSARR174 pKa = 11.84VTHH177 pKa = 6.49AQAKK181 pKa = 7.83DD182 pKa = 3.53TTQKK186 pKa = 11.14AQVLDD191 pKa = 3.44TSIQKK196 pKa = 8.32RR197 pKa = 11.84TSYY200 pKa = 11.03SQFNLGDD207 pKa = 3.51RR208 pKa = 11.84QRR210 pKa = 11.84PLSFTEE216 pKa = 3.48VHH218 pKa = 5.94VPVARR223 pKa = 11.84GNYY226 pKa = 9.5RR227 pKa = 11.84GVTQKK232 pKa = 10.13IVAGPPGGYY241 pKa = 10.15VITGPTGCGKK251 pKa = 8.87STVALLPLFSGKK263 pKa = 10.37SSVLIVEE270 pKa = 4.64PTQANAANIFHH281 pKa = 6.87EE282 pKa = 4.91FSNVLPTLHH291 pKa = 6.66EE292 pKa = 4.68AGVIPWNVPPVEE304 pKa = 4.36FTAPTTRR311 pKa = 11.84EE312 pKa = 3.68GSYY315 pKa = 11.31GPLSVTTTDD324 pKa = 3.44KK325 pKa = 11.02LLEE328 pKa = 4.03YY329 pKa = 10.35FEE331 pKa = 5.0WKK333 pKa = 10.51GSLPKK338 pKa = 9.97VDD340 pKa = 4.17YY341 pKa = 11.21LIIDD345 pKa = 4.2EE346 pKa = 4.48FHH348 pKa = 6.92LPIPSMVQTVEE359 pKa = 3.96LLRR362 pKa = 11.84TFALVPKK369 pKa = 9.42YY370 pKa = 10.51IFVSATAVGYY380 pKa = 9.8SVNPEE385 pKa = 3.75LPKK388 pKa = 10.75AVTQTWGQLPIGQIPSKK405 pKa = 10.61MEE407 pKa = 4.12GSDD410 pKa = 3.47LDD412 pKa = 3.53PRR414 pKa = 11.84RR415 pKa = 11.84WWKK418 pKa = 10.84RR419 pKa = 11.84GDD421 pKa = 3.43GNVAVVAPSVIVAKK435 pKa = 10.55RR436 pKa = 11.84LFNVYY441 pKa = 10.07RR442 pKa = 11.84DD443 pKa = 3.42WQIRR447 pKa = 11.84AFLITRR453 pKa = 11.84EE454 pKa = 4.39TFVSEE459 pKa = 3.95YY460 pKa = 9.96MKK462 pKa = 10.83AATNYY467 pKa = 10.21RR468 pKa = 11.84SMTTFVLEE476 pKa = 4.39PGVEE480 pKa = 4.1AGVTLCIDD488 pKa = 3.48VLII491 pKa = 4.86
Molecular weight: 54.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4565 |
338 |
2128 |
1141.3 |
127.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.375 ± 0.429 | 1.358 ± 0.252 |
6.243 ± 0.649 | 5.608 ± 0.266 |
5.016 ± 0.177 | 6.813 ± 0.229 |
1.752 ± 0.034 | 4.951 ± 0.171 |
5.849 ± 0.222 | 8.105 ± 0.414 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.928 ± 0.17 | 3.264 ± 0.174 |
4.666 ± 0.348 | 3.22 ± 0.221 |
6.396 ± 0.274 | 7.579 ± 0.25 |
6.813 ± 0.623 | 9.332 ± 0.445 |
1.205 ± 0.112 | 3.505 ± 0.165 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
