
Psychromonas sp. psych-6C06
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Psychromonadaceae; Psychromonas; unclassified Psychromonas
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
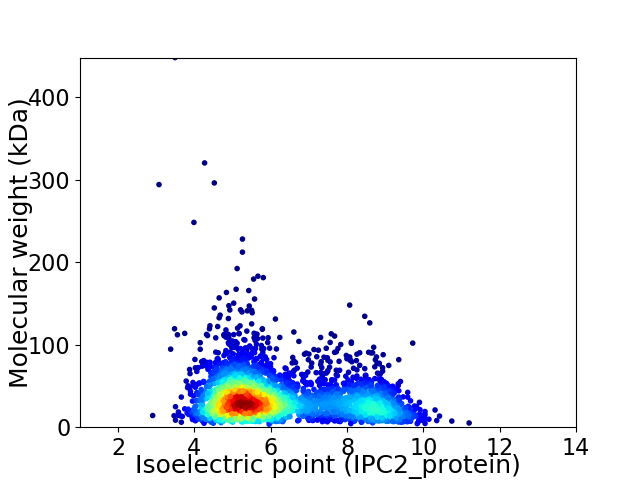
Virtual 2D-PAGE plot for 3200 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
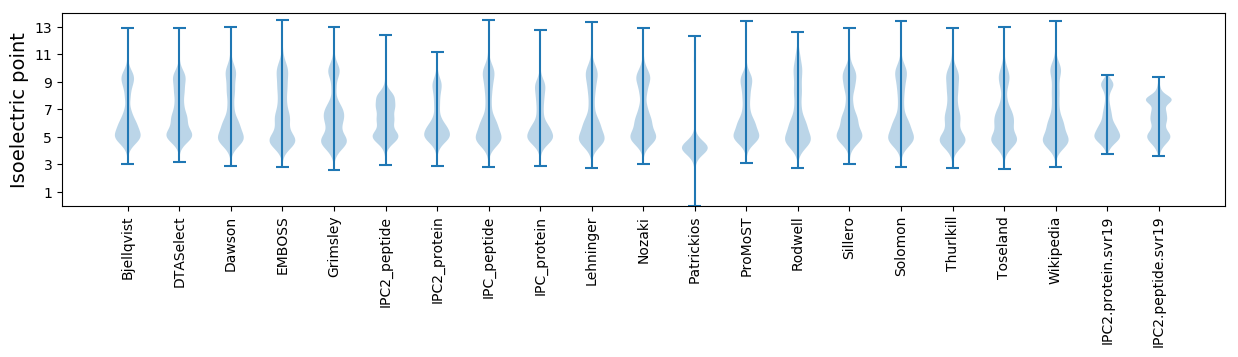
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N0X3Y5|A0A2N0X3Y5_9GAMM Lytic murein transglycosylase OS=Psychromonas sp. psych-6C06 OX=2058089 GN=CW745_01995 PE=4 SV=1
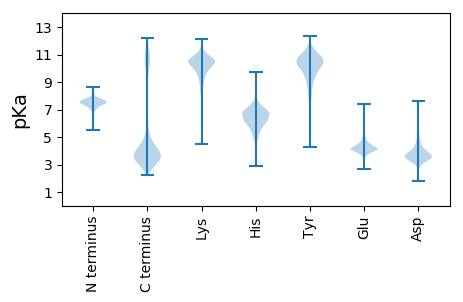
MM1 pKa = 8.2FMMKK5 pKa = 9.9RR6 pKa = 11.84KK7 pKa = 9.66ALLCLPVCLSTLVSMHH23 pKa = 6.43SLANNQDD30 pKa = 2.99ARR32 pKa = 11.84SFAMGGVGVSTSNFLTSSLHH52 pKa = 5.85NPALAAKK59 pKa = 9.52FDD61 pKa = 3.66NSDD64 pKa = 4.1DD65 pKa = 3.56IGFLFPSFAANVNNIADD82 pKa = 3.84TVDD85 pKa = 3.59NIEE88 pKa = 4.45NFSDD92 pKa = 3.86VYY94 pKa = 11.7DD95 pKa = 3.88EE96 pKa = 4.95FKK98 pKa = 11.15SIATPTLDD106 pKa = 4.04DD107 pKa = 3.81AQKK110 pKa = 11.04VVDD113 pKa = 4.77EE114 pKa = 5.05LYY116 pKa = 11.05SLQGDD121 pKa = 3.78PASINAGSQIALAIPNDD138 pKa = 3.32IVAVNLYY145 pKa = 9.62AQAYY149 pKa = 7.47VDD151 pKa = 4.04AMVFADD157 pKa = 5.0IADD160 pKa = 3.99TDD162 pKa = 4.3LEE164 pKa = 4.95ADD166 pKa = 3.9NILNQDD172 pKa = 3.98LNSNALTMGVLVSEE186 pKa = 5.18FGVTLAKK193 pKa = 10.39YY194 pKa = 9.54HH195 pKa = 5.94QFDD198 pKa = 3.92NSTLYY203 pKa = 11.02YY204 pKa = 10.71GLTPKK209 pKa = 10.4YY210 pKa = 9.28QTVDD214 pKa = 3.46TINFVSNIDD223 pKa = 3.91NVDD226 pKa = 3.91FDD228 pKa = 5.05DD229 pKa = 4.53WDD231 pKa = 3.64NDD233 pKa = 3.63SYY235 pKa = 11.83QNSEE239 pKa = 3.62GNMNVDD245 pKa = 4.25LGIAYY250 pKa = 7.87QHH252 pKa = 5.94QNGFGLGLAAKK263 pKa = 9.79NLLKK267 pKa = 10.4QSYY270 pKa = 6.46EE271 pKa = 4.05TEE273 pKa = 4.45SISGVQGHH281 pKa = 5.66YY282 pKa = 10.39EE283 pKa = 3.71ITPVYY288 pKa = 9.15TLGGHH293 pKa = 5.38YY294 pKa = 10.11EE295 pKa = 4.05SSFVIAALDD304 pKa = 3.38IQLNEE309 pKa = 3.97SEE311 pKa = 5.15GYY313 pKa = 10.52SKK315 pKa = 9.95VTGINTNFNSDD326 pKa = 2.87NDD328 pKa = 3.68NRR330 pKa = 11.84QFASLGVEE338 pKa = 4.2FVPMNWIKK346 pKa = 10.75LRR348 pKa = 11.84AGYY351 pKa = 10.38RR352 pKa = 11.84SDD354 pKa = 4.58LSDD357 pKa = 5.15NIDD360 pKa = 3.6DD361 pKa = 5.41SITAGIGFSVVNIFHH376 pKa = 7.23IDD378 pKa = 3.43LSGSYY383 pKa = 10.63SDD385 pKa = 4.26NNNMGAAAQTSFTFF399 pKa = 3.85
MM1 pKa = 8.2FMMKK5 pKa = 9.9RR6 pKa = 11.84KK7 pKa = 9.66ALLCLPVCLSTLVSMHH23 pKa = 6.43SLANNQDD30 pKa = 2.99ARR32 pKa = 11.84SFAMGGVGVSTSNFLTSSLHH52 pKa = 5.85NPALAAKK59 pKa = 9.52FDD61 pKa = 3.66NSDD64 pKa = 4.1DD65 pKa = 3.56IGFLFPSFAANVNNIADD82 pKa = 3.84TVDD85 pKa = 3.59NIEE88 pKa = 4.45NFSDD92 pKa = 3.86VYY94 pKa = 11.7DD95 pKa = 3.88EE96 pKa = 4.95FKK98 pKa = 11.15SIATPTLDD106 pKa = 4.04DD107 pKa = 3.81AQKK110 pKa = 11.04VVDD113 pKa = 4.77EE114 pKa = 5.05LYY116 pKa = 11.05SLQGDD121 pKa = 3.78PASINAGSQIALAIPNDD138 pKa = 3.32IVAVNLYY145 pKa = 9.62AQAYY149 pKa = 7.47VDD151 pKa = 4.04AMVFADD157 pKa = 5.0IADD160 pKa = 3.99TDD162 pKa = 4.3LEE164 pKa = 4.95ADD166 pKa = 3.9NILNQDD172 pKa = 3.98LNSNALTMGVLVSEE186 pKa = 5.18FGVTLAKK193 pKa = 10.39YY194 pKa = 9.54HH195 pKa = 5.94QFDD198 pKa = 3.92NSTLYY203 pKa = 11.02YY204 pKa = 10.71GLTPKK209 pKa = 10.4YY210 pKa = 9.28QTVDD214 pKa = 3.46TINFVSNIDD223 pKa = 3.91NVDD226 pKa = 3.91FDD228 pKa = 5.05DD229 pKa = 4.53WDD231 pKa = 3.64NDD233 pKa = 3.63SYY235 pKa = 11.83QNSEE239 pKa = 3.62GNMNVDD245 pKa = 4.25LGIAYY250 pKa = 7.87QHH252 pKa = 5.94QNGFGLGLAAKK263 pKa = 9.79NLLKK267 pKa = 10.4QSYY270 pKa = 6.46EE271 pKa = 4.05TEE273 pKa = 4.45SISGVQGHH281 pKa = 5.66YY282 pKa = 10.39EE283 pKa = 3.71ITPVYY288 pKa = 9.15TLGGHH293 pKa = 5.38YY294 pKa = 10.11EE295 pKa = 4.05SSFVIAALDD304 pKa = 3.38IQLNEE309 pKa = 3.97SEE311 pKa = 5.15GYY313 pKa = 10.52SKK315 pKa = 9.95VTGINTNFNSDD326 pKa = 2.87NDD328 pKa = 3.68NRR330 pKa = 11.84QFASLGVEE338 pKa = 4.2FVPMNWIKK346 pKa = 10.75LRR348 pKa = 11.84AGYY351 pKa = 10.38RR352 pKa = 11.84SDD354 pKa = 4.58LSDD357 pKa = 5.15NIDD360 pKa = 3.6DD361 pKa = 5.41SITAGIGFSVVNIFHH376 pKa = 7.23IDD378 pKa = 3.43LSGSYY383 pKa = 10.63SDD385 pKa = 4.26NNNMGAAAQTSFTFF399 pKa = 3.85
Molecular weight: 43.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N0WXC0|A0A2N0WXC0_9GAMM Uncharacterized protein OS=Psychromonas sp. psych-6C06 OX=2058089 GN=CW745_13085 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84SHH16 pKa = 6.31GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.2NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84SHH16 pKa = 6.31GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.2NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1074385 |
34 |
4285 |
335.7 |
37.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.215 ± 0.046 | 1.046 ± 0.014 |
5.503 ± 0.052 | 6.121 ± 0.041 |
4.346 ± 0.031 | 6.356 ± 0.047 |
2.165 ± 0.026 | 7.111 ± 0.037 |
5.899 ± 0.047 | 10.709 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.404 ± 0.027 | 4.843 ± 0.039 |
3.598 ± 0.028 | 4.801 ± 0.041 |
3.888 ± 0.032 | 6.841 ± 0.041 |
5.414 ± 0.045 | 6.521 ± 0.035 |
1.107 ± 0.016 | 3.109 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
