
Psychromonas sp. RZ22
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Psychromonadaceae; Psychromonas; unclassified Psychromonas
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
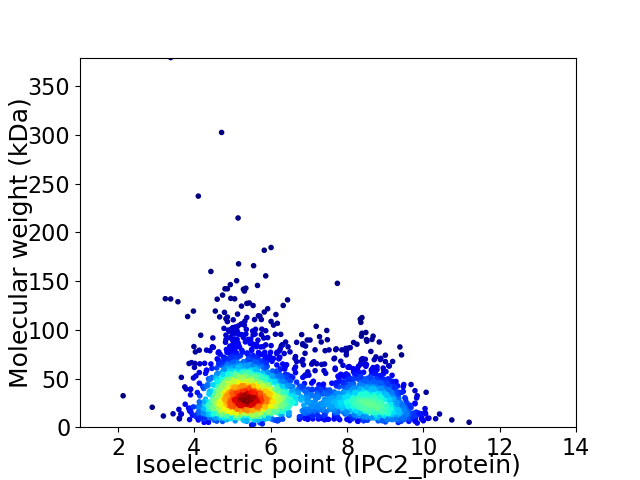
Virtual 2D-PAGE plot for 2647 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y7ZS86|A0A4Y7ZS86_9GAMM Uncharacterized protein OS=Psychromonas sp. RZ22 OX=2555643 GN=E2R68_10975 PE=4 SV=1
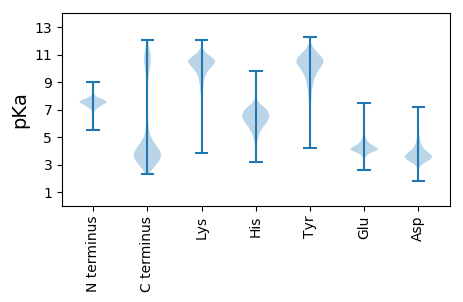
MM1 pKa = 7.16SFKK4 pKa = 11.08YY5 pKa = 10.18NALALITASLLVTACGSSSSSSDD28 pKa = 3.19NGNTTEE34 pKa = 5.84DD35 pKa = 4.02GEE37 pKa = 4.59QEE39 pKa = 3.65ITSNYY44 pKa = 6.75EE45 pKa = 3.91TVEE48 pKa = 3.86LDD50 pKa = 3.31ATDD53 pKa = 3.69GSTAVQLDD61 pKa = 4.19LASGAIVTDD70 pKa = 4.44DD71 pKa = 3.61SWHH74 pKa = 6.51LAYY77 pKa = 10.19QKK79 pKa = 11.29YY80 pKa = 10.64LGFKK84 pKa = 10.62LNGGSSGDD92 pKa = 3.71AGVSGCIAHH101 pKa = 7.2EE102 pKa = 4.01YY103 pKa = 11.01SSIFDD108 pKa = 3.44STGASIEE115 pKa = 4.55DD116 pKa = 3.8EE117 pKa = 4.57FKK119 pKa = 11.26LLTVSSTEE127 pKa = 3.89DD128 pKa = 3.12SFVAVTADD136 pKa = 2.98SCTDD140 pKa = 3.77FVEE143 pKa = 5.42DD144 pKa = 3.87SVATFIEE151 pKa = 4.41TEE153 pKa = 3.72EE154 pKa = 4.32WLDD157 pKa = 3.26ADD159 pKa = 4.18YY160 pKa = 11.53SAGAPVYY167 pKa = 9.93GAKK170 pKa = 10.37AGNGWIIRR178 pKa = 11.84SADD181 pKa = 2.9GATYY185 pKa = 10.73GRR187 pKa = 11.84VSVNSVEE194 pKa = 4.53VVFGASTTRR203 pKa = 11.84KK204 pKa = 9.62LVFNSEE210 pKa = 3.99LWNGSVFEE218 pKa = 4.22TAQLSPEE225 pKa = 4.37LDD227 pKa = 3.61FSEE230 pKa = 5.07DD231 pKa = 3.09QVYY234 pKa = 9.95WDD236 pKa = 5.03LEE238 pKa = 4.55TNSLVSATDD247 pKa = 3.13DD248 pKa = 3.5WEE250 pKa = 4.58LSVSVNGRR258 pKa = 11.84DD259 pKa = 3.47YY260 pKa = 11.08PLQINGGASGTGQAGVGAVLSDD282 pKa = 4.08DD283 pKa = 4.18LASVTDD289 pKa = 3.93PTDD292 pKa = 2.99TAQVYY297 pKa = 10.14KK298 pKa = 10.71YY299 pKa = 10.39FADD302 pKa = 3.77TASGVLSGPGTYY314 pKa = 10.61GPLQYY319 pKa = 11.08AVAGGHH325 pKa = 6.39LMWPTFTTYY334 pKa = 10.92LIKK337 pKa = 10.63DD338 pKa = 3.31AQDD341 pKa = 3.18RR342 pKa = 11.84LFKK345 pKa = 10.77VQVVSNYY352 pKa = 9.22GANGNSASGNLVLRR366 pKa = 11.84YY367 pKa = 10.07EE368 pKa = 4.32EE369 pKa = 4.71LNN371 pKa = 3.55
MM1 pKa = 7.16SFKK4 pKa = 11.08YY5 pKa = 10.18NALALITASLLVTACGSSSSSSDD28 pKa = 3.19NGNTTEE34 pKa = 5.84DD35 pKa = 4.02GEE37 pKa = 4.59QEE39 pKa = 3.65ITSNYY44 pKa = 6.75EE45 pKa = 3.91TVEE48 pKa = 3.86LDD50 pKa = 3.31ATDD53 pKa = 3.69GSTAVQLDD61 pKa = 4.19LASGAIVTDD70 pKa = 4.44DD71 pKa = 3.61SWHH74 pKa = 6.51LAYY77 pKa = 10.19QKK79 pKa = 11.29YY80 pKa = 10.64LGFKK84 pKa = 10.62LNGGSSGDD92 pKa = 3.71AGVSGCIAHH101 pKa = 7.2EE102 pKa = 4.01YY103 pKa = 11.01SSIFDD108 pKa = 3.44STGASIEE115 pKa = 4.55DD116 pKa = 3.8EE117 pKa = 4.57FKK119 pKa = 11.26LLTVSSTEE127 pKa = 3.89DD128 pKa = 3.12SFVAVTADD136 pKa = 2.98SCTDD140 pKa = 3.77FVEE143 pKa = 5.42DD144 pKa = 3.87SVATFIEE151 pKa = 4.41TEE153 pKa = 3.72EE154 pKa = 4.32WLDD157 pKa = 3.26ADD159 pKa = 4.18YY160 pKa = 11.53SAGAPVYY167 pKa = 9.93GAKK170 pKa = 10.37AGNGWIIRR178 pKa = 11.84SADD181 pKa = 2.9GATYY185 pKa = 10.73GRR187 pKa = 11.84VSVNSVEE194 pKa = 4.53VVFGASTTRR203 pKa = 11.84KK204 pKa = 9.62LVFNSEE210 pKa = 3.99LWNGSVFEE218 pKa = 4.22TAQLSPEE225 pKa = 4.37LDD227 pKa = 3.61FSEE230 pKa = 5.07DD231 pKa = 3.09QVYY234 pKa = 9.95WDD236 pKa = 5.03LEE238 pKa = 4.55TNSLVSATDD247 pKa = 3.13DD248 pKa = 3.5WEE250 pKa = 4.58LSVSVNGRR258 pKa = 11.84DD259 pKa = 3.47YY260 pKa = 11.08PLQINGGASGTGQAGVGAVLSDD282 pKa = 4.08DD283 pKa = 4.18LASVTDD289 pKa = 3.93PTDD292 pKa = 2.99TAQVYY297 pKa = 10.14KK298 pKa = 10.71YY299 pKa = 10.39FADD302 pKa = 3.77TASGVLSGPGTYY314 pKa = 10.61GPLQYY319 pKa = 11.08AVAGGHH325 pKa = 6.39LMWPTFTTYY334 pKa = 10.92LIKK337 pKa = 10.63DD338 pKa = 3.31AQDD341 pKa = 3.18RR342 pKa = 11.84LFKK345 pKa = 10.77VQVVSNYY352 pKa = 9.22GANGNSASGNLVLRR366 pKa = 11.84YY367 pKa = 10.07EE368 pKa = 4.32EE369 pKa = 4.71LNN371 pKa = 3.55
Molecular weight: 39.21 kDa
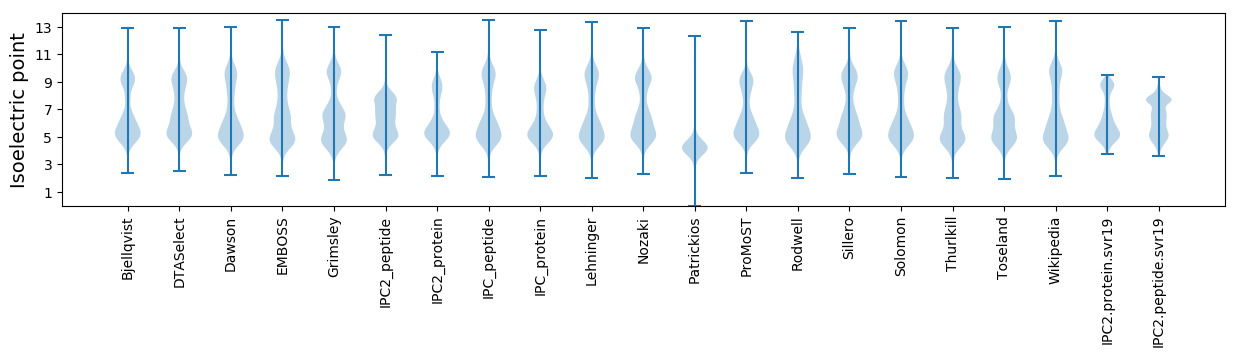
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y7ZVI7|A0A4Y7ZVI7_9GAMM LysR family transcriptional regulator OS=Psychromonas sp. RZ22 OX=2555643 GN=E2R68_09655 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNTKK11 pKa = 10.0RR12 pKa = 11.84KK13 pKa = 8.73RR14 pKa = 11.84SHH16 pKa = 6.16GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.18NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNTKK11 pKa = 10.0RR12 pKa = 11.84KK13 pKa = 8.73RR14 pKa = 11.84SHH16 pKa = 6.16GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.18NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
880803 |
19 |
3594 |
332.8 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.065 ± 0.051 | 1.047 ± 0.016 |
5.433 ± 0.045 | 6.105 ± 0.045 |
4.354 ± 0.035 | 6.349 ± 0.043 |
2.192 ± 0.027 | 7.36 ± 0.048 |
6.14 ± 0.049 | 10.671 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.402 ± 0.023 | 4.807 ± 0.038 |
3.599 ± 0.025 | 4.605 ± 0.044 |
3.821 ± 0.037 | 6.686 ± 0.04 |
5.55 ± 0.043 | 6.581 ± 0.044 |
1.1 ± 0.017 | 3.133 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
