
Lachnospiraceae bacterium 9_1_43BFAA
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
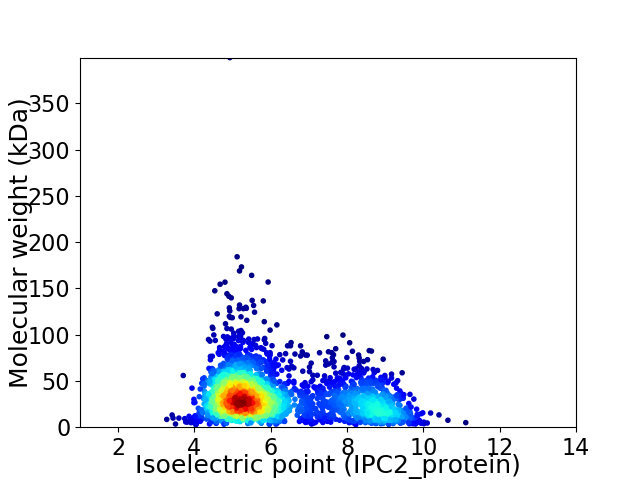
Virtual 2D-PAGE plot for 2717 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F3AMA3|F3AMA3_9FIRM PS_pyruv_trans domain-containing protein OS=Lachnospiraceae bacterium 9_1_43BFAA OX=658088 GN=HMPREF0987_01577 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 9.93IAKK5 pKa = 8.09KK6 pKa = 8.09TAAFLLVLFLGIQCMPLTALAADD29 pKa = 4.12GTLSFSDD36 pKa = 3.69PTAKK40 pKa = 10.39AGEE43 pKa = 4.37TIEE46 pKa = 4.14VTVKK50 pKa = 10.39INTAEE55 pKa = 4.18GPIGDD60 pKa = 4.01GQVQVTYY67 pKa = 10.78DD68 pKa = 3.36PTYY71 pKa = 10.52LEE73 pKa = 4.49FVSGVNATGEE83 pKa = 4.25AGTITLSASGNGIDD97 pKa = 5.3SEE99 pKa = 4.61LAYY102 pKa = 11.49SMTFKK107 pKa = 11.06ALAEE111 pKa = 4.48GEE113 pKa = 4.45TTLQAVGYY121 pKa = 6.33TAYY124 pKa = 9.76TFGGEE129 pKa = 4.21EE130 pKa = 4.32LNLTLGTSAVKK141 pKa = 10.21IEE143 pKa = 4.93AGDD146 pKa = 3.83GTQPAAAQQPGTQTMIEE163 pKa = 4.33LDD165 pKa = 3.35GKK167 pKa = 10.55QYY169 pKa = 10.87FIADD173 pKa = 5.41DD174 pKa = 3.76IPEE177 pKa = 4.08ALIPEE182 pKa = 5.19GYY184 pKa = 9.53EE185 pKa = 3.95KK186 pKa = 10.93SQLTYY191 pKa = 9.99EE192 pKa = 4.92GSPHH196 pKa = 6.81TILVHH201 pKa = 5.9TGSGQKK207 pKa = 9.39MIYY210 pKa = 10.67LMDD213 pKa = 3.72TQGEE217 pKa = 4.75GNLFLYY223 pKa = 10.53DD224 pKa = 3.73SEE226 pKa = 4.61TDD228 pKa = 3.12HH229 pKa = 7.18FSAFEE234 pKa = 4.5KK235 pKa = 10.25IDD237 pKa = 3.99LSSDD241 pKa = 3.48SYY243 pKa = 11.85ILITDD248 pKa = 4.03DD249 pKa = 4.85KK250 pKa = 10.07EE251 pKa = 4.36SANIPEE257 pKa = 5.03DD258 pKa = 3.85YY259 pKa = 10.91EE260 pKa = 4.09EE261 pKa = 4.07TTIQINGKK269 pKa = 8.42VFPAWQNVVDD279 pKa = 4.09TKK281 pKa = 11.2LADD284 pKa = 3.54FCLVYY289 pKa = 10.81AVDD292 pKa = 3.76NAGKK296 pKa = 10.22KK297 pKa = 10.15GLYY300 pKa = 9.92QYY302 pKa = 11.59DD303 pKa = 4.06STQKK307 pKa = 8.86TYY309 pKa = 10.86QRR311 pKa = 11.84FLPPTAEE318 pKa = 4.31KK319 pKa = 10.18EE320 pKa = 4.24KK321 pKa = 10.73QDD323 pKa = 3.18GSVANKK329 pKa = 7.72VTTFVEE335 pKa = 4.27EE336 pKa = 4.1NLPKK340 pKa = 10.46IMVGAWGVFLMMLLLIVILAVKK362 pKa = 9.86LRR364 pKa = 11.84HH365 pKa = 6.14RR366 pKa = 11.84NEE368 pKa = 4.12EE369 pKa = 3.66LDD371 pKa = 4.09DD372 pKa = 4.83YY373 pKa = 11.62YY374 pKa = 11.44DD375 pKa = 3.56QEE377 pKa = 4.23YY378 pKa = 11.24DD379 pKa = 3.65EE380 pKa = 5.02NPRR383 pKa = 11.84AARR386 pKa = 11.84GFSSEE391 pKa = 5.54DD392 pKa = 3.38YY393 pKa = 11.1DD394 pKa = 5.18EE395 pKa = 6.52SDD397 pKa = 4.85DD398 pKa = 5.26DD399 pKa = 5.42DD400 pKa = 5.18YY401 pKa = 11.81DD402 pKa = 4.19YY403 pKa = 11.92EE404 pKa = 4.18EE405 pKa = 6.18DD406 pKa = 3.98YY407 pKa = 11.64DD408 pKa = 4.79EE409 pKa = 6.65SDD411 pKa = 4.67DD412 pKa = 5.26DD413 pKa = 5.42DD414 pKa = 5.18YY415 pKa = 11.81DD416 pKa = 4.19YY417 pKa = 11.92EE418 pKa = 4.15EE419 pKa = 6.12DD420 pKa = 3.77YY421 pKa = 11.62DD422 pKa = 4.17EE423 pKa = 6.19SEE425 pKa = 4.36EE426 pKa = 4.91DD427 pKa = 3.45DD428 pKa = 4.0YY429 pKa = 11.9AYY431 pKa = 10.47EE432 pKa = 4.15DD433 pKa = 4.73AYY435 pKa = 11.13DD436 pKa = 4.03EE437 pKa = 5.65SEE439 pKa = 5.05DD440 pKa = 5.8DD441 pKa = 4.26YY442 pKa = 11.96DD443 pKa = 5.53DD444 pKa = 5.99DD445 pKa = 4.69YY446 pKa = 11.8DD447 pKa = 4.34YY448 pKa = 11.95EE449 pKa = 4.2EE450 pKa = 5.84DD451 pKa = 4.09YY452 pKa = 11.21EE453 pKa = 5.69SEE455 pKa = 4.16DD456 pKa = 4.92AYY458 pKa = 11.25DD459 pKa = 5.31DD460 pKa = 4.47LLDD463 pKa = 4.93YY464 pKa = 11.55YY465 pKa = 11.17DD466 pKa = 6.09DD467 pKa = 5.61DD468 pKa = 6.36DD469 pKa = 6.7DD470 pKa = 5.98YY471 pKa = 11.85EE472 pKa = 4.6LEE474 pKa = 4.74RR475 pKa = 11.84EE476 pKa = 4.36ILDD479 pKa = 3.54EE480 pKa = 5.53DD481 pKa = 3.64EE482 pKa = 5.29GFFDD486 pKa = 4.62KK487 pKa = 10.95RR488 pKa = 11.84SGKK491 pKa = 9.8KK492 pKa = 9.97RR493 pKa = 11.84GIDD496 pKa = 3.79LII498 pKa = 4.39
MM1 pKa = 7.64KK2 pKa = 9.93IAKK5 pKa = 8.09KK6 pKa = 8.09TAAFLLVLFLGIQCMPLTALAADD29 pKa = 4.12GTLSFSDD36 pKa = 3.69PTAKK40 pKa = 10.39AGEE43 pKa = 4.37TIEE46 pKa = 4.14VTVKK50 pKa = 10.39INTAEE55 pKa = 4.18GPIGDD60 pKa = 4.01GQVQVTYY67 pKa = 10.78DD68 pKa = 3.36PTYY71 pKa = 10.52LEE73 pKa = 4.49FVSGVNATGEE83 pKa = 4.25AGTITLSASGNGIDD97 pKa = 5.3SEE99 pKa = 4.61LAYY102 pKa = 11.49SMTFKK107 pKa = 11.06ALAEE111 pKa = 4.48GEE113 pKa = 4.45TTLQAVGYY121 pKa = 6.33TAYY124 pKa = 9.76TFGGEE129 pKa = 4.21EE130 pKa = 4.32LNLTLGTSAVKK141 pKa = 10.21IEE143 pKa = 4.93AGDD146 pKa = 3.83GTQPAAAQQPGTQTMIEE163 pKa = 4.33LDD165 pKa = 3.35GKK167 pKa = 10.55QYY169 pKa = 10.87FIADD173 pKa = 5.41DD174 pKa = 3.76IPEE177 pKa = 4.08ALIPEE182 pKa = 5.19GYY184 pKa = 9.53EE185 pKa = 3.95KK186 pKa = 10.93SQLTYY191 pKa = 9.99EE192 pKa = 4.92GSPHH196 pKa = 6.81TILVHH201 pKa = 5.9TGSGQKK207 pKa = 9.39MIYY210 pKa = 10.67LMDD213 pKa = 3.72TQGEE217 pKa = 4.75GNLFLYY223 pKa = 10.53DD224 pKa = 3.73SEE226 pKa = 4.61TDD228 pKa = 3.12HH229 pKa = 7.18FSAFEE234 pKa = 4.5KK235 pKa = 10.25IDD237 pKa = 3.99LSSDD241 pKa = 3.48SYY243 pKa = 11.85ILITDD248 pKa = 4.03DD249 pKa = 4.85KK250 pKa = 10.07EE251 pKa = 4.36SANIPEE257 pKa = 5.03DD258 pKa = 3.85YY259 pKa = 10.91EE260 pKa = 4.09EE261 pKa = 4.07TTIQINGKK269 pKa = 8.42VFPAWQNVVDD279 pKa = 4.09TKK281 pKa = 11.2LADD284 pKa = 3.54FCLVYY289 pKa = 10.81AVDD292 pKa = 3.76NAGKK296 pKa = 10.22KK297 pKa = 10.15GLYY300 pKa = 9.92QYY302 pKa = 11.59DD303 pKa = 4.06STQKK307 pKa = 8.86TYY309 pKa = 10.86QRR311 pKa = 11.84FLPPTAEE318 pKa = 4.31KK319 pKa = 10.18EE320 pKa = 4.24KK321 pKa = 10.73QDD323 pKa = 3.18GSVANKK329 pKa = 7.72VTTFVEE335 pKa = 4.27EE336 pKa = 4.1NLPKK340 pKa = 10.46IMVGAWGVFLMMLLLIVILAVKK362 pKa = 9.86LRR364 pKa = 11.84HH365 pKa = 6.14RR366 pKa = 11.84NEE368 pKa = 4.12EE369 pKa = 3.66LDD371 pKa = 4.09DD372 pKa = 4.83YY373 pKa = 11.62YY374 pKa = 11.44DD375 pKa = 3.56QEE377 pKa = 4.23YY378 pKa = 11.24DD379 pKa = 3.65EE380 pKa = 5.02NPRR383 pKa = 11.84AARR386 pKa = 11.84GFSSEE391 pKa = 5.54DD392 pKa = 3.38YY393 pKa = 11.1DD394 pKa = 5.18EE395 pKa = 6.52SDD397 pKa = 4.85DD398 pKa = 5.26DD399 pKa = 5.42DD400 pKa = 5.18YY401 pKa = 11.81DD402 pKa = 4.19YY403 pKa = 11.92EE404 pKa = 4.18EE405 pKa = 6.18DD406 pKa = 3.98YY407 pKa = 11.64DD408 pKa = 4.79EE409 pKa = 6.65SDD411 pKa = 4.67DD412 pKa = 5.26DD413 pKa = 5.42DD414 pKa = 5.18YY415 pKa = 11.81DD416 pKa = 4.19YY417 pKa = 11.92EE418 pKa = 4.15EE419 pKa = 6.12DD420 pKa = 3.77YY421 pKa = 11.62DD422 pKa = 4.17EE423 pKa = 6.19SEE425 pKa = 4.36EE426 pKa = 4.91DD427 pKa = 3.45DD428 pKa = 4.0YY429 pKa = 11.9AYY431 pKa = 10.47EE432 pKa = 4.15DD433 pKa = 4.73AYY435 pKa = 11.13DD436 pKa = 4.03EE437 pKa = 5.65SEE439 pKa = 5.05DD440 pKa = 5.8DD441 pKa = 4.26YY442 pKa = 11.96DD443 pKa = 5.53DD444 pKa = 5.99DD445 pKa = 4.69YY446 pKa = 11.8DD447 pKa = 4.34YY448 pKa = 11.95EE449 pKa = 4.2EE450 pKa = 5.84DD451 pKa = 4.09YY452 pKa = 11.21EE453 pKa = 5.69SEE455 pKa = 4.16DD456 pKa = 4.92AYY458 pKa = 11.25DD459 pKa = 5.31DD460 pKa = 4.47LLDD463 pKa = 4.93YY464 pKa = 11.55YY465 pKa = 11.17DD466 pKa = 6.09DD467 pKa = 5.61DD468 pKa = 6.36DD469 pKa = 6.7DD470 pKa = 5.98YY471 pKa = 11.85EE472 pKa = 4.6LEE474 pKa = 4.74RR475 pKa = 11.84EE476 pKa = 4.36ILDD479 pKa = 3.54EE480 pKa = 5.53DD481 pKa = 3.64EE482 pKa = 5.29GFFDD486 pKa = 4.62KK487 pKa = 10.95RR488 pKa = 11.84SGKK491 pKa = 9.8KK492 pKa = 9.97RR493 pKa = 11.84GIDD496 pKa = 3.79LII498 pKa = 4.39
Molecular weight: 55.86 kDa
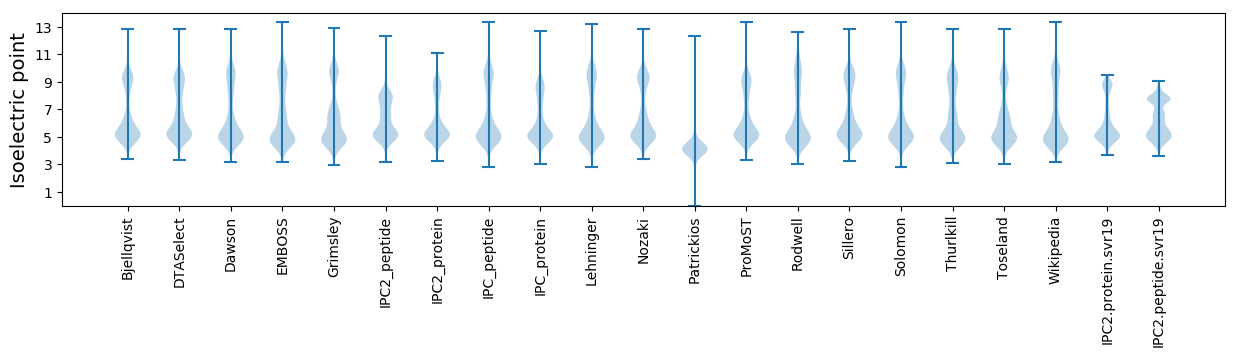
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F3APH0|F3APH0_9FIRM Fumerase_C domain-containing protein OS=Lachnospiraceae bacterium 9_1_43BFAA OX=658088 GN=HMPREF0987_02344 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
Molecular weight: 4.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
842039 |
22 |
3638 |
309.9 |
34.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.124 ± 0.046 | 1.494 ± 0.019 |
5.073 ± 0.035 | 8.558 ± 0.062 |
4.199 ± 0.029 | 7.032 ± 0.042 |
1.775 ± 0.022 | 7.446 ± 0.045 |
7.277 ± 0.043 | 9.033 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.151 ± 0.024 | 4.014 ± 0.032 |
3.166 ± 0.025 | 3.491 ± 0.031 |
4.44 ± 0.033 | 5.461 ± 0.037 |
5.253 ± 0.034 | 6.98 ± 0.04 |
0.913 ± 0.016 | 4.119 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
