
Alces alces faeces associated smacovirus MP78
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus alecas1
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
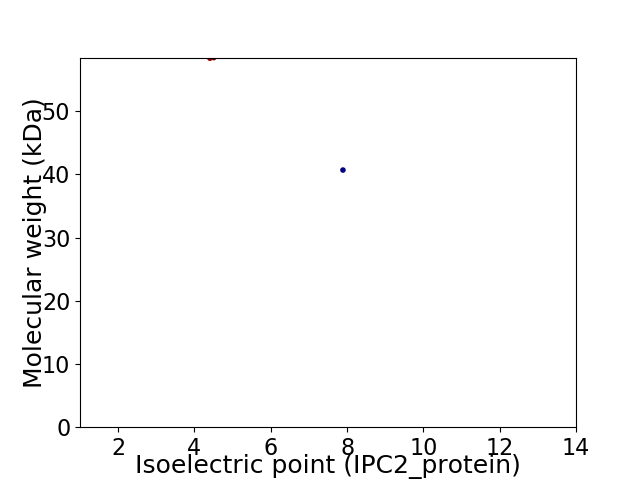
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CL51|A0A2Z5CL51_9VIRU Capsid protein OS=Alces alces faeces associated smacovirus MP78 OX=2219142 PE=4 SV=1
MM1 pKa = 6.48TTNKK5 pKa = 10.07AFASYY10 pKa = 10.94HH11 pKa = 5.92EE12 pKa = 5.38IIDD15 pKa = 3.8LHH17 pKa = 5.65TEE19 pKa = 3.76SDD21 pKa = 3.57KK22 pKa = 11.68VGVIGIHH29 pKa = 5.99TPTDD33 pKa = 3.73YY34 pKa = 10.68TPHH37 pKa = 7.21RR38 pKa = 11.84MFGGYY43 pKa = 9.63FKK45 pKa = 10.6QFKK48 pKa = 9.05KK49 pKa = 10.66FKK51 pKa = 10.49YY52 pKa = 8.83LGCSISLVPAARR64 pKa = 11.84LPADD68 pKa = 4.3PSQVSYY74 pKa = 11.05GAGEE78 pKa = 4.1PPIDD82 pKa = 4.96PRR84 pKa = 11.84DD85 pKa = 3.68LLNPIMFHH93 pKa = 6.19GCHH96 pKa = 6.32GNDD99 pKa = 4.09LGNVLNRR106 pKa = 11.84LYY108 pKa = 10.18TLNSDD113 pKa = 3.73GVSGGFATNIQEE125 pKa = 4.19SDD127 pKa = 3.5SVLKK131 pKa = 10.82FEE133 pKa = 5.77EE134 pKa = 4.35ITNSYY139 pKa = 10.6YY140 pKa = 10.76DD141 pKa = 4.52DD142 pKa = 3.99NLLEE146 pKa = 4.02NLYY149 pKa = 10.94YY150 pKa = 10.57KK151 pKa = 10.91ALTDD155 pKa = 3.59NTWRR159 pKa = 11.84KK160 pKa = 8.79AHH162 pKa = 5.84PQRR165 pKa = 11.84GFRR168 pKa = 11.84KK169 pKa = 9.87GGLRR173 pKa = 11.84PLIYY177 pKa = 11.04SMATNVQIMNPNEE190 pKa = 3.84FSTEE194 pKa = 3.62RR195 pKa = 11.84LAGSNGHH202 pKa = 6.97LSITSPINTGMPEE215 pKa = 3.79DD216 pKa = 3.89FAFSVSGSSGRR227 pKa = 11.84FSRR230 pKa = 11.84GVIDD234 pKa = 5.36DD235 pKa = 3.93GVKK238 pKa = 10.42VFTPHH243 pKa = 5.83LTGLGWLDD251 pKa = 3.42TRR253 pKa = 11.84SVITQGVEE261 pKa = 4.19SDD263 pKa = 3.06WTTLSDD269 pKa = 3.29TDD271 pKa = 5.19AIASAQDD278 pKa = 3.49SIVNKK283 pKa = 10.09EE284 pKa = 4.23VVTTLPKK291 pKa = 10.24IFMGVILLPPAYY303 pKa = 8.68KK304 pKa = 9.84TEE306 pKa = 3.6QYY308 pKa = 10.98YY309 pKa = 11.1RR310 pKa = 11.84MIINHH315 pKa = 6.47RR316 pKa = 11.84FGFKK320 pKa = 10.29DD321 pKa = 3.4FRR323 pKa = 11.84GISFANDD330 pKa = 3.18QLVTPAYY337 pKa = 10.77ANFNDD342 pKa = 4.47SDD344 pKa = 4.28YY345 pKa = 11.02EE346 pKa = 4.24YY347 pKa = 11.02NPPEE351 pKa = 4.91DD352 pKa = 4.52PDD354 pKa = 4.0SGEE357 pKa = 4.04GDD359 pKa = 3.78SPSLSGSEE367 pKa = 3.98YY368 pKa = 10.73LVSDD372 pKa = 3.92LQQYY376 pKa = 6.57FTSYY380 pKa = 11.03VGMGTWKK387 pKa = 10.68LDD389 pKa = 3.27TQYY392 pKa = 10.86PQTVYY397 pKa = 9.2EE398 pKa = 4.4TLTEE402 pKa = 4.07YY403 pKa = 11.15DD404 pKa = 3.51SGEE407 pKa = 4.21STLSVSFPVQNQSDD421 pKa = 4.28GNSTGWGYY429 pKa = 10.96NGNWSCSAFWYY440 pKa = 10.12LDD442 pKa = 3.04NGFVYY447 pKa = 10.31LGIMLCEE454 pKa = 3.61EE455 pKa = 4.25SLYY458 pKa = 11.13GFYY461 pKa = 10.61LWQNSEE467 pKa = 3.64WTFQNWSDD475 pKa = 3.79SLDD478 pKa = 4.34LIGSGSITQLQADD491 pKa = 3.96VAFAGLVNSNSSTVDD506 pKa = 3.66DD507 pKa = 4.5LNQYY511 pKa = 8.12QIKK514 pKa = 10.1SLSGNIGDD522 pKa = 4.31VDD524 pKa = 3.71NAA526 pKa = 4.01
MM1 pKa = 6.48TTNKK5 pKa = 10.07AFASYY10 pKa = 10.94HH11 pKa = 5.92EE12 pKa = 5.38IIDD15 pKa = 3.8LHH17 pKa = 5.65TEE19 pKa = 3.76SDD21 pKa = 3.57KK22 pKa = 11.68VGVIGIHH29 pKa = 5.99TPTDD33 pKa = 3.73YY34 pKa = 10.68TPHH37 pKa = 7.21RR38 pKa = 11.84MFGGYY43 pKa = 9.63FKK45 pKa = 10.6QFKK48 pKa = 9.05KK49 pKa = 10.66FKK51 pKa = 10.49YY52 pKa = 8.83LGCSISLVPAARR64 pKa = 11.84LPADD68 pKa = 4.3PSQVSYY74 pKa = 11.05GAGEE78 pKa = 4.1PPIDD82 pKa = 4.96PRR84 pKa = 11.84DD85 pKa = 3.68LLNPIMFHH93 pKa = 6.19GCHH96 pKa = 6.32GNDD99 pKa = 4.09LGNVLNRR106 pKa = 11.84LYY108 pKa = 10.18TLNSDD113 pKa = 3.73GVSGGFATNIQEE125 pKa = 4.19SDD127 pKa = 3.5SVLKK131 pKa = 10.82FEE133 pKa = 5.77EE134 pKa = 4.35ITNSYY139 pKa = 10.6YY140 pKa = 10.76DD141 pKa = 4.52DD142 pKa = 3.99NLLEE146 pKa = 4.02NLYY149 pKa = 10.94YY150 pKa = 10.57KK151 pKa = 10.91ALTDD155 pKa = 3.59NTWRR159 pKa = 11.84KK160 pKa = 8.79AHH162 pKa = 5.84PQRR165 pKa = 11.84GFRR168 pKa = 11.84KK169 pKa = 9.87GGLRR173 pKa = 11.84PLIYY177 pKa = 11.04SMATNVQIMNPNEE190 pKa = 3.84FSTEE194 pKa = 3.62RR195 pKa = 11.84LAGSNGHH202 pKa = 6.97LSITSPINTGMPEE215 pKa = 3.79DD216 pKa = 3.89FAFSVSGSSGRR227 pKa = 11.84FSRR230 pKa = 11.84GVIDD234 pKa = 5.36DD235 pKa = 3.93GVKK238 pKa = 10.42VFTPHH243 pKa = 5.83LTGLGWLDD251 pKa = 3.42TRR253 pKa = 11.84SVITQGVEE261 pKa = 4.19SDD263 pKa = 3.06WTTLSDD269 pKa = 3.29TDD271 pKa = 5.19AIASAQDD278 pKa = 3.49SIVNKK283 pKa = 10.09EE284 pKa = 4.23VVTTLPKK291 pKa = 10.24IFMGVILLPPAYY303 pKa = 8.68KK304 pKa = 9.84TEE306 pKa = 3.6QYY308 pKa = 10.98YY309 pKa = 11.1RR310 pKa = 11.84MIINHH315 pKa = 6.47RR316 pKa = 11.84FGFKK320 pKa = 10.29DD321 pKa = 3.4FRR323 pKa = 11.84GISFANDD330 pKa = 3.18QLVTPAYY337 pKa = 10.77ANFNDD342 pKa = 4.47SDD344 pKa = 4.28YY345 pKa = 11.02EE346 pKa = 4.24YY347 pKa = 11.02NPPEE351 pKa = 4.91DD352 pKa = 4.52PDD354 pKa = 4.0SGEE357 pKa = 4.04GDD359 pKa = 3.78SPSLSGSEE367 pKa = 3.98YY368 pKa = 10.73LVSDD372 pKa = 3.92LQQYY376 pKa = 6.57FTSYY380 pKa = 11.03VGMGTWKK387 pKa = 10.68LDD389 pKa = 3.27TQYY392 pKa = 10.86PQTVYY397 pKa = 9.2EE398 pKa = 4.4TLTEE402 pKa = 4.07YY403 pKa = 11.15DD404 pKa = 3.51SGEE407 pKa = 4.21STLSVSFPVQNQSDD421 pKa = 4.28GNSTGWGYY429 pKa = 10.96NGNWSCSAFWYY440 pKa = 10.12LDD442 pKa = 3.04NGFVYY447 pKa = 10.31LGIMLCEE454 pKa = 3.61EE455 pKa = 4.25SLYY458 pKa = 11.13GFYY461 pKa = 10.61LWQNSEE467 pKa = 3.64WTFQNWSDD475 pKa = 3.79SLDD478 pKa = 4.34LIGSGSITQLQADD491 pKa = 3.96VAFAGLVNSNSSTVDD506 pKa = 3.66DD507 pKa = 4.5LNQYY511 pKa = 8.12QIKK514 pKa = 10.1SLSGNIGDD522 pKa = 4.31VDD524 pKa = 3.71NAA526 pKa = 4.01
Molecular weight: 58.4 kDa
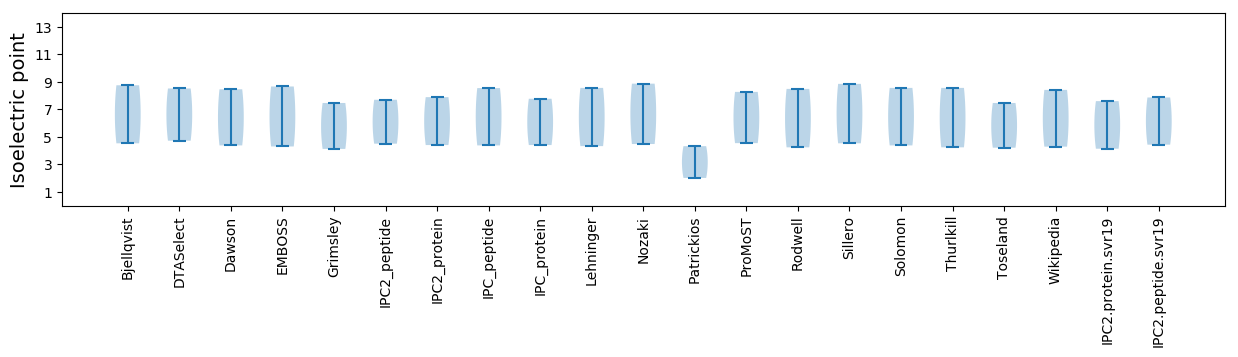
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CL51|A0A2Z5CL51_9VIRU Capsid protein OS=Alces alces faeces associated smacovirus MP78 OX=2219142 PE=4 SV=1
MM1 pKa = 7.38QRR3 pKa = 11.84TNKK6 pKa = 9.31NRR8 pKa = 11.84SRR10 pKa = 11.84QVQDD14 pKa = 3.05QSAGIISTFSCDD26 pKa = 3.19NIYY29 pKa = 10.48VHH31 pKa = 7.0NAFCKK36 pKa = 8.57TYY38 pKa = 10.21AYY40 pKa = 8.5HH41 pKa = 6.7TFLSLLYY48 pKa = 9.38EE49 pKa = 4.21YY50 pKa = 10.51RR51 pKa = 11.84SNALSTLVSRR61 pKa = 11.84MLTKK65 pKa = 9.85ICYY68 pKa = 9.55LRR70 pKa = 11.84YY71 pKa = 9.24QRR73 pKa = 11.84CGRR76 pKa = 11.84QEE78 pKa = 3.54AHH80 pKa = 6.15MVKK83 pKa = 8.42TWMLTVPRR91 pKa = 11.84NGDD94 pKa = 2.72WCTTQSVCRR103 pKa = 11.84MIQEE107 pKa = 4.07NDD109 pKa = 3.17VKK111 pKa = 10.72KK112 pKa = 7.72WTIGFEE118 pKa = 4.07EE119 pKa = 4.82GKK121 pKa = 10.19NGYY124 pKa = 8.04RR125 pKa = 11.84HH126 pKa = 4.49YY127 pKa = 10.66QIRR130 pKa = 11.84LVSSNHH136 pKa = 7.1DD137 pKa = 3.3FFEE140 pKa = 4.05WCKK143 pKa = 10.72INLPTAHH150 pKa = 6.89IEE152 pKa = 4.08EE153 pKa = 4.45ATEE156 pKa = 3.66EE157 pKa = 4.14SGNYY161 pKa = 7.47EE162 pKa = 3.78RR163 pKa = 11.84KK164 pKa = 10.02SGNFLSSDD172 pKa = 3.29DD173 pKa = 3.73TAEE176 pKa = 3.67IRR178 pKa = 11.84QVRR181 pKa = 11.84FGVLRR186 pKa = 11.84EE187 pKa = 4.09SQKK190 pKa = 11.1KK191 pKa = 9.5ILEE194 pKa = 4.23EE195 pKa = 4.18VNDD198 pKa = 3.63QGDD201 pKa = 3.76RR202 pKa = 11.84EE203 pKa = 4.06IDD205 pKa = 3.63VYY207 pKa = 11.2FDD209 pKa = 3.37PRR211 pKa = 11.84GNNGKK216 pKa = 7.55TWLTIHH222 pKa = 6.92LYY224 pKa = 9.71EE225 pKa = 5.01RR226 pKa = 11.84GRR228 pKa = 11.84ALVVPRR234 pKa = 11.84ANTTAEE240 pKa = 4.09KK241 pKa = 10.3LSAYY245 pKa = 9.35ICSSYY250 pKa = 11.14NGEE253 pKa = 4.12EE254 pKa = 4.01YY255 pKa = 10.95VIIDD259 pKa = 3.56IPRR262 pKa = 11.84SRR264 pKa = 11.84KK265 pKa = 7.6ITPEE269 pKa = 3.18IYY271 pKa = 10.16EE272 pKa = 4.69AIEE275 pKa = 4.26EE276 pKa = 4.33IKK278 pKa = 10.88DD279 pKa = 3.42GLVFDD284 pKa = 4.33SRR286 pKa = 11.84YY287 pKa = 9.92SGRR290 pKa = 11.84SRR292 pKa = 11.84NIRR295 pKa = 11.84GVKK298 pKa = 10.34LIIFTNKK305 pKa = 10.14RR306 pKa = 11.84LDD308 pKa = 3.57LTKK311 pKa = 10.74LSNDD315 pKa = 2.76RR316 pKa = 11.84WRR318 pKa = 11.84LHH320 pKa = 7.11AISTTTKK327 pKa = 10.25RR328 pKa = 11.84PMKK331 pKa = 9.47TDD333 pKa = 3.55YY334 pKa = 11.14VPSDD338 pKa = 4.8DD339 pKa = 4.67DD340 pKa = 3.87TAPDD344 pKa = 3.52GAKK347 pKa = 10.49AVDD350 pKa = 3.31
MM1 pKa = 7.38QRR3 pKa = 11.84TNKK6 pKa = 9.31NRR8 pKa = 11.84SRR10 pKa = 11.84QVQDD14 pKa = 3.05QSAGIISTFSCDD26 pKa = 3.19NIYY29 pKa = 10.48VHH31 pKa = 7.0NAFCKK36 pKa = 8.57TYY38 pKa = 10.21AYY40 pKa = 8.5HH41 pKa = 6.7TFLSLLYY48 pKa = 9.38EE49 pKa = 4.21YY50 pKa = 10.51RR51 pKa = 11.84SNALSTLVSRR61 pKa = 11.84MLTKK65 pKa = 9.85ICYY68 pKa = 9.55LRR70 pKa = 11.84YY71 pKa = 9.24QRR73 pKa = 11.84CGRR76 pKa = 11.84QEE78 pKa = 3.54AHH80 pKa = 6.15MVKK83 pKa = 8.42TWMLTVPRR91 pKa = 11.84NGDD94 pKa = 2.72WCTTQSVCRR103 pKa = 11.84MIQEE107 pKa = 4.07NDD109 pKa = 3.17VKK111 pKa = 10.72KK112 pKa = 7.72WTIGFEE118 pKa = 4.07EE119 pKa = 4.82GKK121 pKa = 10.19NGYY124 pKa = 8.04RR125 pKa = 11.84HH126 pKa = 4.49YY127 pKa = 10.66QIRR130 pKa = 11.84LVSSNHH136 pKa = 7.1DD137 pKa = 3.3FFEE140 pKa = 4.05WCKK143 pKa = 10.72INLPTAHH150 pKa = 6.89IEE152 pKa = 4.08EE153 pKa = 4.45ATEE156 pKa = 3.66EE157 pKa = 4.14SGNYY161 pKa = 7.47EE162 pKa = 3.78RR163 pKa = 11.84KK164 pKa = 10.02SGNFLSSDD172 pKa = 3.29DD173 pKa = 3.73TAEE176 pKa = 3.67IRR178 pKa = 11.84QVRR181 pKa = 11.84FGVLRR186 pKa = 11.84EE187 pKa = 4.09SQKK190 pKa = 11.1KK191 pKa = 9.5ILEE194 pKa = 4.23EE195 pKa = 4.18VNDD198 pKa = 3.63QGDD201 pKa = 3.76RR202 pKa = 11.84EE203 pKa = 4.06IDD205 pKa = 3.63VYY207 pKa = 11.2FDD209 pKa = 3.37PRR211 pKa = 11.84GNNGKK216 pKa = 7.55TWLTIHH222 pKa = 6.92LYY224 pKa = 9.71EE225 pKa = 5.01RR226 pKa = 11.84GRR228 pKa = 11.84ALVVPRR234 pKa = 11.84ANTTAEE240 pKa = 4.09KK241 pKa = 10.3LSAYY245 pKa = 9.35ICSSYY250 pKa = 11.14NGEE253 pKa = 4.12EE254 pKa = 4.01YY255 pKa = 10.95VIIDD259 pKa = 3.56IPRR262 pKa = 11.84SRR264 pKa = 11.84KK265 pKa = 7.6ITPEE269 pKa = 3.18IYY271 pKa = 10.16EE272 pKa = 4.69AIEE275 pKa = 4.26EE276 pKa = 4.33IKK278 pKa = 10.88DD279 pKa = 3.42GLVFDD284 pKa = 4.33SRR286 pKa = 11.84YY287 pKa = 9.92SGRR290 pKa = 11.84SRR292 pKa = 11.84NIRR295 pKa = 11.84GVKK298 pKa = 10.34LIIFTNKK305 pKa = 10.14RR306 pKa = 11.84LDD308 pKa = 3.57LTKK311 pKa = 10.74LSNDD315 pKa = 2.76RR316 pKa = 11.84WRR318 pKa = 11.84LHH320 pKa = 7.11AISTTTKK327 pKa = 10.25RR328 pKa = 11.84PMKK331 pKa = 9.47TDD333 pKa = 3.55YY334 pKa = 11.14VPSDD338 pKa = 4.8DD339 pKa = 4.67DD340 pKa = 3.87TAPDD344 pKa = 3.52GAKK347 pKa = 10.49AVDD350 pKa = 3.31
Molecular weight: 40.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
876 |
350 |
526 |
438.0 |
49.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.68 ± 0.103 | 1.37 ± 0.535 |
6.963 ± 0.396 | 5.365 ± 0.871 |
4.338 ± 0.698 | 7.42 ± 1.329 |
2.055 ± 0.135 | 5.936 ± 0.704 |
4.338 ± 0.97 | 7.648 ± 0.629 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.826 ± 0.065 | 6.164 ± 0.263 |
3.881 ± 0.765 | 3.767 ± 0.198 |
5.251 ± 2.105 | 9.132 ± 0.995 |
7.192 ± 0.138 | 5.479 ± 0.03 |
1.826 ± 0.065 | 5.365 ± 0.13 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
