
Azoarcus sp. CIB
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Zoogloeaceae; Azoarcus; unclassified Azoarcus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
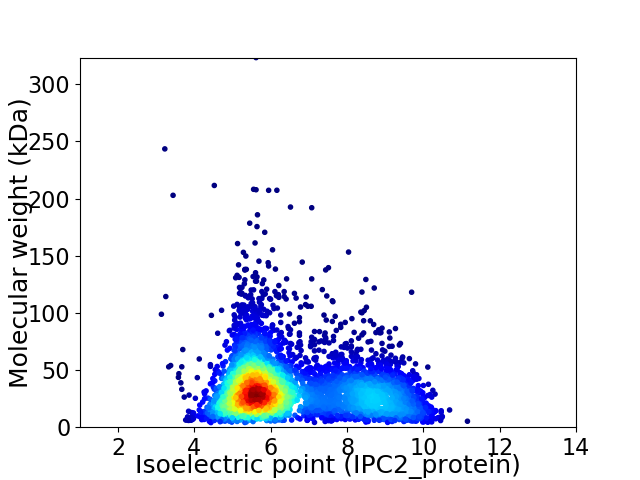
Virtual 2D-PAGE plot for 4693 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1J5Y1|A0A0K1J5Y1_9RHOO EamA domain-containing protein OS=Azoarcus sp. CIB OX=198107 GN=AzCIB_2068 PE=4 SV=1
MM1 pKa = 7.66ALASLSTQQIIDD13 pKa = 3.56QLDD16 pKa = 3.53SGFHH20 pKa = 5.3WVGSTITYY28 pKa = 8.88AFPVIAADD36 pKa = 5.31LFWTDD41 pKa = 3.26EE42 pKa = 4.25GSSFRR47 pKa = 11.84PLDD50 pKa = 3.62ATQQAYY56 pKa = 10.66AEE58 pKa = 4.37VALASWDD65 pKa = 3.74EE66 pKa = 4.44LIAPSFAEE74 pKa = 4.22VSGSASDD81 pKa = 3.88VEE83 pKa = 5.51FGLTLNADD91 pKa = 3.5YY92 pKa = 11.34AHH94 pKa = 7.64AYY96 pKa = 8.27FPSYY100 pKa = 10.22GSVWFRR106 pKa = 11.84SGDD109 pKa = 3.27ASLADD114 pKa = 4.26PEE116 pKa = 4.48VGSYY120 pKa = 11.08GYY122 pKa = 8.62LTYY125 pKa = 10.84VHH127 pKa = 6.65EE128 pKa = 4.78VGHH131 pKa = 6.75ALGLDD136 pKa = 3.22HH137 pKa = 6.92MGDD140 pKa = 3.84YY141 pKa = 10.72NGYY144 pKa = 9.51APEE147 pKa = 4.2PTSYY151 pKa = 10.85QDD153 pKa = 3.39STVLSVMSYY162 pKa = 9.84FGPSYY167 pKa = 10.76GSWSAGTGQVAWADD181 pKa = 3.54WVGSDD186 pKa = 4.03GRR188 pKa = 11.84LYY190 pKa = 10.56QPQTPMLDD198 pKa = 3.24DD199 pKa = 3.52VLAIQSIYY207 pKa = 10.66GADD210 pKa = 3.44RR211 pKa = 11.84TTRR214 pKa = 11.84PGDD217 pKa = 3.25TTYY220 pKa = 11.45GFGSNVSGAMAEE232 pKa = 3.99ILDD235 pKa = 4.29FARR238 pKa = 11.84NAHH241 pKa = 6.37PVLILYY247 pKa = 8.73DD248 pKa = 3.57AGGNDD253 pKa = 3.96TLNLSGWSSGSTVDD267 pKa = 3.77IAPGAYY273 pKa = 9.71SSCNDD278 pKa = 3.31MTNNIAVAYY287 pKa = 8.36STDD290 pKa = 3.27IEE292 pKa = 4.42NVVCGDD298 pKa = 3.55GSDD301 pKa = 4.18SISGNTLANRR311 pKa = 11.84LDD313 pKa = 4.0GGGGNDD319 pKa = 3.77TIYY322 pKa = 10.7GQSGDD327 pKa = 4.5DD328 pKa = 3.56EE329 pKa = 4.87LLGGGGNDD337 pKa = 3.87HH338 pKa = 7.41LDD340 pKa = 3.66GGTGIDD346 pKa = 3.76WATLGADD353 pKa = 3.57LGSYY357 pKa = 10.27AIIYY361 pKa = 8.8DD362 pKa = 3.59AATGIFTFQNQSTGVDD378 pKa = 3.75TITGTEE384 pKa = 4.0YY385 pKa = 10.54FRR387 pKa = 11.84FANDD391 pKa = 3.07VTRR394 pKa = 11.84SANEE398 pKa = 3.99LIGGQRR404 pKa = 11.84AAPVAGITVSGTGRR418 pKa = 11.84ANVLTGTDD426 pKa = 3.64ADD428 pKa = 4.08DD429 pKa = 3.55QLFGLGGADD438 pKa = 3.56SLDD441 pKa = 3.35GGFGHH446 pKa = 7.74DD447 pKa = 4.39LLDD450 pKa = 4.19GGRR453 pKa = 11.84SADD456 pKa = 3.85RR457 pKa = 11.84LAGGNGDD464 pKa = 3.51DD465 pKa = 4.43TYY467 pKa = 11.99VVDD470 pKa = 3.9NRR472 pKa = 11.84NDD474 pKa = 3.1IVIEE478 pKa = 4.3SNGLLGGIDD487 pKa = 3.69TVRR490 pKa = 11.84STVSFALGADD500 pKa = 4.14LEE502 pKa = 4.55HH503 pKa = 6.62LTLTGRR509 pKa = 11.84ARR511 pKa = 11.84ANGAGNDD518 pKa = 3.89GNNVIVGNAGLNTLDD533 pKa = 4.35GRR535 pKa = 11.84SGDD538 pKa = 3.81DD539 pKa = 3.35TLIGAGGRR547 pKa = 11.84DD548 pKa = 3.61ILSGGAGHH556 pKa = 7.56DD557 pKa = 3.29VFRR560 pKa = 11.84FDD562 pKa = 5.9AISDD566 pKa = 3.74TSATARR572 pKa = 11.84KK573 pKa = 9.89ADD575 pKa = 4.08MISDD579 pKa = 3.92FAIGEE584 pKa = 4.24DD585 pKa = 4.37LLDD588 pKa = 5.11LSLIDD593 pKa = 5.08ANLLTAEE600 pKa = 4.46NDD602 pKa = 3.3AFDD605 pKa = 3.98ATFVAAGSAFYY616 pKa = 10.99APGQLQFAGGILYY629 pKa = 10.92GNVDD633 pKa = 3.83ADD635 pKa = 4.14AKK637 pKa = 10.96PEE639 pKa = 3.69FAINVGQLALSHH651 pKa = 6.82GDD653 pKa = 3.44LVLL656 pKa = 4.83
MM1 pKa = 7.66ALASLSTQQIIDD13 pKa = 3.56QLDD16 pKa = 3.53SGFHH20 pKa = 5.3WVGSTITYY28 pKa = 8.88AFPVIAADD36 pKa = 5.31LFWTDD41 pKa = 3.26EE42 pKa = 4.25GSSFRR47 pKa = 11.84PLDD50 pKa = 3.62ATQQAYY56 pKa = 10.66AEE58 pKa = 4.37VALASWDD65 pKa = 3.74EE66 pKa = 4.44LIAPSFAEE74 pKa = 4.22VSGSASDD81 pKa = 3.88VEE83 pKa = 5.51FGLTLNADD91 pKa = 3.5YY92 pKa = 11.34AHH94 pKa = 7.64AYY96 pKa = 8.27FPSYY100 pKa = 10.22GSVWFRR106 pKa = 11.84SGDD109 pKa = 3.27ASLADD114 pKa = 4.26PEE116 pKa = 4.48VGSYY120 pKa = 11.08GYY122 pKa = 8.62LTYY125 pKa = 10.84VHH127 pKa = 6.65EE128 pKa = 4.78VGHH131 pKa = 6.75ALGLDD136 pKa = 3.22HH137 pKa = 6.92MGDD140 pKa = 3.84YY141 pKa = 10.72NGYY144 pKa = 9.51APEE147 pKa = 4.2PTSYY151 pKa = 10.85QDD153 pKa = 3.39STVLSVMSYY162 pKa = 9.84FGPSYY167 pKa = 10.76GSWSAGTGQVAWADD181 pKa = 3.54WVGSDD186 pKa = 4.03GRR188 pKa = 11.84LYY190 pKa = 10.56QPQTPMLDD198 pKa = 3.24DD199 pKa = 3.52VLAIQSIYY207 pKa = 10.66GADD210 pKa = 3.44RR211 pKa = 11.84TTRR214 pKa = 11.84PGDD217 pKa = 3.25TTYY220 pKa = 11.45GFGSNVSGAMAEE232 pKa = 3.99ILDD235 pKa = 4.29FARR238 pKa = 11.84NAHH241 pKa = 6.37PVLILYY247 pKa = 8.73DD248 pKa = 3.57AGGNDD253 pKa = 3.96TLNLSGWSSGSTVDD267 pKa = 3.77IAPGAYY273 pKa = 9.71SSCNDD278 pKa = 3.31MTNNIAVAYY287 pKa = 8.36STDD290 pKa = 3.27IEE292 pKa = 4.42NVVCGDD298 pKa = 3.55GSDD301 pKa = 4.18SISGNTLANRR311 pKa = 11.84LDD313 pKa = 4.0GGGGNDD319 pKa = 3.77TIYY322 pKa = 10.7GQSGDD327 pKa = 4.5DD328 pKa = 3.56EE329 pKa = 4.87LLGGGGNDD337 pKa = 3.87HH338 pKa = 7.41LDD340 pKa = 3.66GGTGIDD346 pKa = 3.76WATLGADD353 pKa = 3.57LGSYY357 pKa = 10.27AIIYY361 pKa = 8.8DD362 pKa = 3.59AATGIFTFQNQSTGVDD378 pKa = 3.75TITGTEE384 pKa = 4.0YY385 pKa = 10.54FRR387 pKa = 11.84FANDD391 pKa = 3.07VTRR394 pKa = 11.84SANEE398 pKa = 3.99LIGGQRR404 pKa = 11.84AAPVAGITVSGTGRR418 pKa = 11.84ANVLTGTDD426 pKa = 3.64ADD428 pKa = 4.08DD429 pKa = 3.55QLFGLGGADD438 pKa = 3.56SLDD441 pKa = 3.35GGFGHH446 pKa = 7.74DD447 pKa = 4.39LLDD450 pKa = 4.19GGRR453 pKa = 11.84SADD456 pKa = 3.85RR457 pKa = 11.84LAGGNGDD464 pKa = 3.51DD465 pKa = 4.43TYY467 pKa = 11.99VVDD470 pKa = 3.9NRR472 pKa = 11.84NDD474 pKa = 3.1IVIEE478 pKa = 4.3SNGLLGGIDD487 pKa = 3.69TVRR490 pKa = 11.84STVSFALGADD500 pKa = 4.14LEE502 pKa = 4.55HH503 pKa = 6.62LTLTGRR509 pKa = 11.84ARR511 pKa = 11.84ANGAGNDD518 pKa = 3.89GNNVIVGNAGLNTLDD533 pKa = 4.35GRR535 pKa = 11.84SGDD538 pKa = 3.81DD539 pKa = 3.35TLIGAGGRR547 pKa = 11.84DD548 pKa = 3.61ILSGGAGHH556 pKa = 7.56DD557 pKa = 3.29VFRR560 pKa = 11.84FDD562 pKa = 5.9AISDD566 pKa = 3.74TSATARR572 pKa = 11.84KK573 pKa = 9.89ADD575 pKa = 4.08MISDD579 pKa = 3.92FAIGEE584 pKa = 4.24DD585 pKa = 4.37LLDD588 pKa = 5.11LSLIDD593 pKa = 5.08ANLLTAEE600 pKa = 4.46NDD602 pKa = 3.3AFDD605 pKa = 3.98ATFVAAGSAFYY616 pKa = 10.99APGQLQFAGGILYY629 pKa = 10.92GNVDD633 pKa = 3.83ADD635 pKa = 4.14AKK637 pKa = 10.96PEE639 pKa = 3.69FAINVGQLALSHH651 pKa = 6.82GDD653 pKa = 3.44LVLL656 pKa = 4.83
Molecular weight: 67.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1JD67|A0A0K1JD67_9RHOO Putative IIA component of sugar transport PTS system OS=Azoarcus sp. CIB OX=198107 GN=AzCIB_4649 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1569040 |
37 |
2929 |
334.3 |
36.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.581 ± 0.046 | 1.033 ± 0.012 |
5.629 ± 0.03 | 5.878 ± 0.033 |
3.621 ± 0.024 | 8.419 ± 0.037 |
2.26 ± 0.016 | 4.646 ± 0.027 |
3.095 ± 0.03 | 10.583 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.385 ± 0.017 | 2.583 ± 0.021 |
5.122 ± 0.029 | 3.27 ± 0.019 |
7.567 ± 0.036 | 5.062 ± 0.023 |
4.972 ± 0.029 | 7.649 ± 0.033 |
1.383 ± 0.016 | 2.263 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
