
Shuangao sobemo-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.05
Get precalculated fractions of proteins
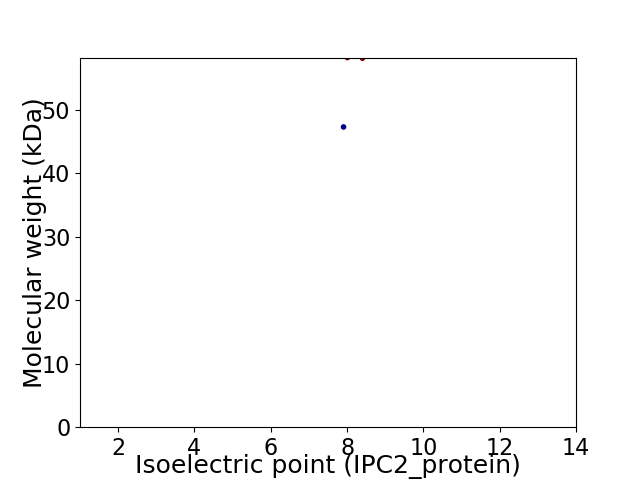
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEF6|A0A1L3KEF6_9VIRU RdRP_1 domain-containing protein OS=Shuangao sobemo-like virus 2 OX=1923475 PE=4 SV=1
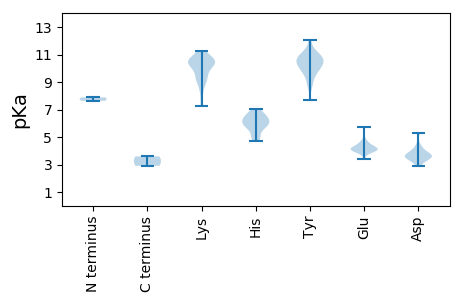
MM1 pKa = 7.88LSMMRR6 pKa = 11.84NSVQVVPMAASALAGAAAYY25 pKa = 9.52QVGKK29 pKa = 10.06IASQAYY35 pKa = 9.83DD36 pKa = 3.5SLKK39 pKa = 10.82SPPAPPTFYY48 pKa = 10.9EE49 pKa = 3.96QLLEE53 pKa = 4.1MGLEE57 pKa = 4.12RR58 pKa = 11.84KK59 pKa = 8.28KK60 pKa = 9.46TIAVALFGAAAVGFWACKK78 pKa = 7.56NRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84VLLKK87 pKa = 10.27PLSQEE92 pKa = 3.99SLVAGSAEE100 pKa = 4.12MPALEE105 pKa = 4.94PACQVAIAFEE115 pKa = 4.7SPDD118 pKa = 3.27GTLVKK123 pKa = 10.43VGSGVRR129 pKa = 11.84GLVGCNHH136 pKa = 5.75VLFTAAHH143 pKa = 5.88NLSFEE148 pKa = 4.11QPLWLIKK155 pKa = 10.38GSKK158 pKa = 7.26QVRR161 pKa = 11.84LGDD164 pKa = 3.65YY165 pKa = 10.95SPIPLAADD173 pKa = 3.47AALLLVPEE181 pKa = 4.62KK182 pKa = 10.48TFGLLGVKK190 pKa = 9.07VANFSPLPGKK200 pKa = 10.17SVAVSVTGLAGKK212 pKa = 8.29GTTGILSPVVNPHH225 pKa = 6.3HH226 pKa = 7.06GIGHH230 pKa = 4.74VTYY233 pKa = 10.52AATTAGGYY241 pKa = 9.83SGAPYY246 pKa = 10.5VSGSQVYY253 pKa = 9.93GIHH256 pKa = 5.91IHH258 pKa = 6.32GGMRR262 pKa = 11.84NGGFEE267 pKa = 3.92ILYY270 pKa = 10.13LYY272 pKa = 10.97ALAKK276 pKa = 10.04IALEE280 pKa = 3.98YY281 pKa = 10.6TEE283 pKa = 5.69EE284 pKa = 4.06DD285 pKa = 3.56TEE287 pKa = 4.41DD288 pKa = 3.55AFMEE292 pKa = 4.29RR293 pKa = 11.84LFNSDD298 pKa = 3.51DD299 pKa = 3.43PDD301 pKa = 4.19YY302 pKa = 10.96YY303 pKa = 10.58VQEE306 pKa = 4.41VGDD309 pKa = 3.83KK310 pKa = 10.9VIVRR314 pKa = 11.84VHH316 pKa = 6.21GNSHH320 pKa = 4.95YY321 pKa = 10.9HH322 pKa = 5.7VVDD325 pKa = 3.59AKK327 pKa = 10.86KK328 pKa = 10.53FRR330 pKa = 11.84DD331 pKa = 3.83YY332 pKa = 10.75EE333 pKa = 4.29ARR335 pKa = 11.84WEE337 pKa = 4.09NADD340 pKa = 3.78YY341 pKa = 11.39SSDD344 pKa = 3.97DD345 pKa = 3.68YY346 pKa = 12.09VEE348 pKa = 4.64EE349 pKa = 4.45CASEE353 pKa = 4.03ALNFQSPGQRR363 pKa = 11.84SRR365 pKa = 11.84AGPSIEE371 pKa = 4.16CLHH374 pKa = 6.33RR375 pKa = 11.84QPQLVTSSCQTSALPEE391 pKa = 4.14SSPSSDD397 pKa = 4.38RR398 pKa = 11.84RR399 pKa = 11.84PQDD402 pKa = 3.19PSNKK406 pKa = 9.32RR407 pKa = 11.84LKK409 pKa = 10.7KK410 pKa = 10.45LINSRR415 pKa = 11.84LRR417 pKa = 11.84RR418 pKa = 11.84LQLKK422 pKa = 9.28TNGTRR427 pKa = 11.84STPSRR432 pKa = 11.84SLVTQVV438 pKa = 2.91
MM1 pKa = 7.88LSMMRR6 pKa = 11.84NSVQVVPMAASALAGAAAYY25 pKa = 9.52QVGKK29 pKa = 10.06IASQAYY35 pKa = 9.83DD36 pKa = 3.5SLKK39 pKa = 10.82SPPAPPTFYY48 pKa = 10.9EE49 pKa = 3.96QLLEE53 pKa = 4.1MGLEE57 pKa = 4.12RR58 pKa = 11.84KK59 pKa = 8.28KK60 pKa = 9.46TIAVALFGAAAVGFWACKK78 pKa = 7.56NRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84VLLKK87 pKa = 10.27PLSQEE92 pKa = 3.99SLVAGSAEE100 pKa = 4.12MPALEE105 pKa = 4.94PACQVAIAFEE115 pKa = 4.7SPDD118 pKa = 3.27GTLVKK123 pKa = 10.43VGSGVRR129 pKa = 11.84GLVGCNHH136 pKa = 5.75VLFTAAHH143 pKa = 5.88NLSFEE148 pKa = 4.11QPLWLIKK155 pKa = 10.38GSKK158 pKa = 7.26QVRR161 pKa = 11.84LGDD164 pKa = 3.65YY165 pKa = 10.95SPIPLAADD173 pKa = 3.47AALLLVPEE181 pKa = 4.62KK182 pKa = 10.48TFGLLGVKK190 pKa = 9.07VANFSPLPGKK200 pKa = 10.17SVAVSVTGLAGKK212 pKa = 8.29GTTGILSPVVNPHH225 pKa = 6.3HH226 pKa = 7.06GIGHH230 pKa = 4.74VTYY233 pKa = 10.52AATTAGGYY241 pKa = 9.83SGAPYY246 pKa = 10.5VSGSQVYY253 pKa = 9.93GIHH256 pKa = 5.91IHH258 pKa = 6.32GGMRR262 pKa = 11.84NGGFEE267 pKa = 3.92ILYY270 pKa = 10.13LYY272 pKa = 10.97ALAKK276 pKa = 10.04IALEE280 pKa = 3.98YY281 pKa = 10.6TEE283 pKa = 5.69EE284 pKa = 4.06DD285 pKa = 3.56TEE287 pKa = 4.41DD288 pKa = 3.55AFMEE292 pKa = 4.29RR293 pKa = 11.84LFNSDD298 pKa = 3.51DD299 pKa = 3.43PDD301 pKa = 4.19YY302 pKa = 10.96YY303 pKa = 10.58VQEE306 pKa = 4.41VGDD309 pKa = 3.83KK310 pKa = 10.9VIVRR314 pKa = 11.84VHH316 pKa = 6.21GNSHH320 pKa = 4.95YY321 pKa = 10.9HH322 pKa = 5.7VVDD325 pKa = 3.59AKK327 pKa = 10.86KK328 pKa = 10.53FRR330 pKa = 11.84DD331 pKa = 3.83YY332 pKa = 10.75EE333 pKa = 4.29ARR335 pKa = 11.84WEE337 pKa = 4.09NADD340 pKa = 3.78YY341 pKa = 11.39SSDD344 pKa = 3.97DD345 pKa = 3.68YY346 pKa = 12.09VEE348 pKa = 4.64EE349 pKa = 4.45CASEE353 pKa = 4.03ALNFQSPGQRR363 pKa = 11.84SRR365 pKa = 11.84AGPSIEE371 pKa = 4.16CLHH374 pKa = 6.33RR375 pKa = 11.84QPQLVTSSCQTSALPEE391 pKa = 4.14SSPSSDD397 pKa = 4.38RR398 pKa = 11.84RR399 pKa = 11.84PQDD402 pKa = 3.19PSNKK406 pKa = 9.32RR407 pKa = 11.84LKK409 pKa = 10.7KK410 pKa = 10.45LINSRR415 pKa = 11.84LRR417 pKa = 11.84RR418 pKa = 11.84LQLKK422 pKa = 9.28TNGTRR427 pKa = 11.84STPSRR432 pKa = 11.84SLVTQVV438 pKa = 2.91
Molecular weight: 47.27 kDa
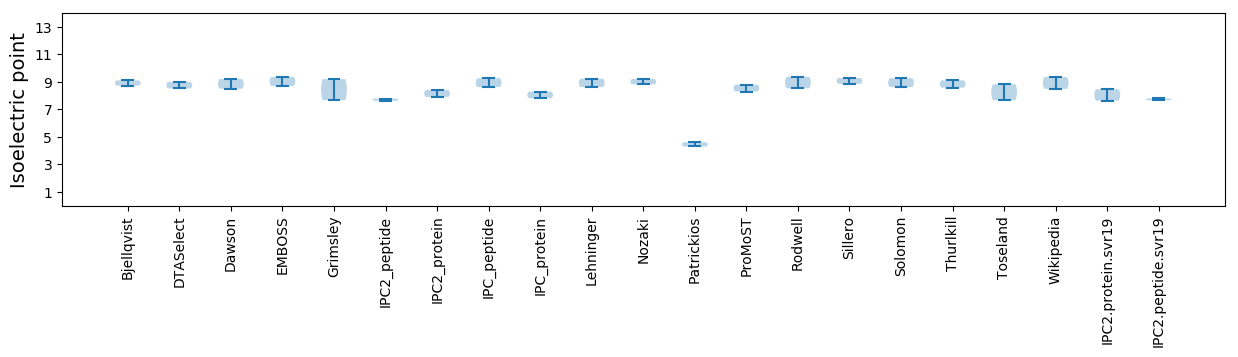
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEF6|A0A1L3KEF6_9VIRU RdRP_1 domain-containing protein OS=Shuangao sobemo-like virus 2 OX=1923475 PE=4 SV=1
MM1 pKa = 7.63CFGSFKK7 pKa = 10.61LPKK10 pKa = 9.68PGTAVPGWTLYY21 pKa = 10.53RR22 pKa = 11.84VLAPTATAGNKK33 pKa = 8.73LVPNEE38 pKa = 3.38RR39 pKa = 11.84VARR42 pKa = 11.84VFAEE46 pKa = 4.32LRR48 pKa = 11.84PTAPGPIKK56 pKa = 10.3QAFEE60 pKa = 4.02EE61 pKa = 4.39ADD63 pKa = 3.73KK64 pKa = 10.39LTAPPTTAKK73 pKa = 10.55DD74 pKa = 3.39QWDD77 pKa = 3.61SFYY80 pKa = 8.83TQSVARR86 pKa = 11.84HH87 pKa = 5.81ASLKK91 pKa = 10.13TYY93 pKa = 10.48VEE95 pKa = 3.77PDD97 pKa = 2.95EE98 pKa = 5.73RR99 pKa = 11.84LFSMILYY106 pKa = 9.61QAEE109 pKa = 4.15QTYY112 pKa = 10.51SSCIRR117 pKa = 11.84KK118 pKa = 9.04LPLGWDD124 pKa = 3.5TRR126 pKa = 11.84SHH128 pKa = 6.63FDD130 pKa = 2.86KK131 pKa = 10.75TLRR134 pKa = 11.84KK135 pKa = 9.41LDD137 pKa = 3.74RR138 pKa = 11.84TSSPGWPLCKK148 pKa = 9.58QASTIGDD155 pKa = 3.58WLYY158 pKa = 10.45PGSILDD164 pKa = 4.32PDD166 pKa = 3.92PMKK169 pKa = 10.98AEE171 pKa = 4.21QLWMMVQQVFEE182 pKa = 4.5LEE184 pKa = 4.06YY185 pKa = 9.88EE186 pKa = 4.15HH187 pKa = 6.8FFKK190 pKa = 11.22VFIKK194 pKa = 10.36PEE196 pKa = 3.69PHH198 pKa = 6.78KK199 pKa = 10.6EE200 pKa = 4.03EE201 pKa = 4.23KK202 pKa = 10.48SLARR206 pKa = 11.84RR207 pKa = 11.84WRR209 pKa = 11.84LIMASSLPVQIAWHH223 pKa = 5.7MAMGHH228 pKa = 5.94LEE230 pKa = 3.94EE231 pKa = 5.35AFLRR235 pKa = 11.84EE236 pKa = 4.16QPFIPLAYY244 pKa = 10.03GEE246 pKa = 4.56PFFAGSWLRR255 pKa = 11.84FKK257 pKa = 10.67EE258 pKa = 4.29SCKK261 pKa = 9.27KK262 pKa = 9.22TKK264 pKa = 9.02ITWATDD270 pKa = 2.87KK271 pKa = 11.19KK272 pKa = 10.39AWDD275 pKa = 3.69WNSPGWVYY283 pKa = 8.89QACRR287 pKa = 11.84EE288 pKa = 4.06LRR290 pKa = 11.84VRR292 pKa = 11.84LTRR295 pKa = 11.84FDD297 pKa = 3.64HH298 pKa = 6.39THH300 pKa = 6.26DD301 pKa = 3.25QQRR304 pKa = 11.84WFRR307 pKa = 11.84VVNWLYY313 pKa = 11.36DD314 pKa = 3.45DD315 pKa = 5.28AYY317 pKa = 10.84RR318 pKa = 11.84SSKK321 pKa = 10.61LLLASGHH328 pKa = 6.08VYY330 pKa = 10.08QQTTAGLMKK339 pKa = 10.37SGLVPTISDD348 pKa = 3.31NSLSQDD354 pKa = 4.47LIDD357 pKa = 4.04MAAQLSVGRR366 pKa = 11.84LPCKK370 pKa = 10.34KK371 pKa = 9.88RR372 pKa = 11.84VTGDD376 pKa = 3.48DD377 pKa = 3.97VIQEE381 pKa = 4.28KK382 pKa = 9.64PADD385 pKa = 3.78PQAYY389 pKa = 7.66IQRR392 pKa = 11.84VQSFGCKK399 pKa = 8.8IKK401 pKa = 10.55QAATKK406 pKa = 10.46LEE408 pKa = 3.97FMGFDD413 pKa = 3.38MDD415 pKa = 5.28GDD417 pKa = 4.14LTPIYY422 pKa = 8.8PQKK425 pKa = 10.67HH426 pKa = 5.21LWNFLHH432 pKa = 6.58QKK434 pKa = 10.02EE435 pKa = 4.69EE436 pKa = 4.06YY437 pKa = 10.13LEE439 pKa = 4.19QVVGAYY445 pKa = 10.23LRR447 pKa = 11.84IYY449 pKa = 10.56ANSPQHH455 pKa = 4.98SQFWRR460 pKa = 11.84AVADD464 pKa = 3.79KK465 pKa = 11.25LNLRR469 pKa = 11.84VMSPSFYY476 pKa = 10.9SFFMNNPEE484 pKa = 3.94AAMKK488 pKa = 10.1LGPVRR493 pKa = 11.84YY494 pKa = 9.36RR495 pKa = 11.84EE496 pKa = 4.08GLLGGAGLVV505 pKa = 3.59
MM1 pKa = 7.63CFGSFKK7 pKa = 10.61LPKK10 pKa = 9.68PGTAVPGWTLYY21 pKa = 10.53RR22 pKa = 11.84VLAPTATAGNKK33 pKa = 8.73LVPNEE38 pKa = 3.38RR39 pKa = 11.84VARR42 pKa = 11.84VFAEE46 pKa = 4.32LRR48 pKa = 11.84PTAPGPIKK56 pKa = 10.3QAFEE60 pKa = 4.02EE61 pKa = 4.39ADD63 pKa = 3.73KK64 pKa = 10.39LTAPPTTAKK73 pKa = 10.55DD74 pKa = 3.39QWDD77 pKa = 3.61SFYY80 pKa = 8.83TQSVARR86 pKa = 11.84HH87 pKa = 5.81ASLKK91 pKa = 10.13TYY93 pKa = 10.48VEE95 pKa = 3.77PDD97 pKa = 2.95EE98 pKa = 5.73RR99 pKa = 11.84LFSMILYY106 pKa = 9.61QAEE109 pKa = 4.15QTYY112 pKa = 10.51SSCIRR117 pKa = 11.84KK118 pKa = 9.04LPLGWDD124 pKa = 3.5TRR126 pKa = 11.84SHH128 pKa = 6.63FDD130 pKa = 2.86KK131 pKa = 10.75TLRR134 pKa = 11.84KK135 pKa = 9.41LDD137 pKa = 3.74RR138 pKa = 11.84TSSPGWPLCKK148 pKa = 9.58QASTIGDD155 pKa = 3.58WLYY158 pKa = 10.45PGSILDD164 pKa = 4.32PDD166 pKa = 3.92PMKK169 pKa = 10.98AEE171 pKa = 4.21QLWMMVQQVFEE182 pKa = 4.5LEE184 pKa = 4.06YY185 pKa = 9.88EE186 pKa = 4.15HH187 pKa = 6.8FFKK190 pKa = 11.22VFIKK194 pKa = 10.36PEE196 pKa = 3.69PHH198 pKa = 6.78KK199 pKa = 10.6EE200 pKa = 4.03EE201 pKa = 4.23KK202 pKa = 10.48SLARR206 pKa = 11.84RR207 pKa = 11.84WRR209 pKa = 11.84LIMASSLPVQIAWHH223 pKa = 5.7MAMGHH228 pKa = 5.94LEE230 pKa = 3.94EE231 pKa = 5.35AFLRR235 pKa = 11.84EE236 pKa = 4.16QPFIPLAYY244 pKa = 10.03GEE246 pKa = 4.56PFFAGSWLRR255 pKa = 11.84FKK257 pKa = 10.67EE258 pKa = 4.29SCKK261 pKa = 9.27KK262 pKa = 9.22TKK264 pKa = 9.02ITWATDD270 pKa = 2.87KK271 pKa = 11.19KK272 pKa = 10.39AWDD275 pKa = 3.69WNSPGWVYY283 pKa = 8.89QACRR287 pKa = 11.84EE288 pKa = 4.06LRR290 pKa = 11.84VRR292 pKa = 11.84LTRR295 pKa = 11.84FDD297 pKa = 3.64HH298 pKa = 6.39THH300 pKa = 6.26DD301 pKa = 3.25QQRR304 pKa = 11.84WFRR307 pKa = 11.84VVNWLYY313 pKa = 11.36DD314 pKa = 3.45DD315 pKa = 5.28AYY317 pKa = 10.84RR318 pKa = 11.84SSKK321 pKa = 10.61LLLASGHH328 pKa = 6.08VYY330 pKa = 10.08QQTTAGLMKK339 pKa = 10.37SGLVPTISDD348 pKa = 3.31NSLSQDD354 pKa = 4.47LIDD357 pKa = 4.04MAAQLSVGRR366 pKa = 11.84LPCKK370 pKa = 10.34KK371 pKa = 9.88RR372 pKa = 11.84VTGDD376 pKa = 3.48DD377 pKa = 3.97VIQEE381 pKa = 4.28KK382 pKa = 9.64PADD385 pKa = 3.78PQAYY389 pKa = 7.66IQRR392 pKa = 11.84VQSFGCKK399 pKa = 8.8IKK401 pKa = 10.55QAATKK406 pKa = 10.46LEE408 pKa = 3.97FMGFDD413 pKa = 3.38MDD415 pKa = 5.28GDD417 pKa = 4.14LTPIYY422 pKa = 8.8PQKK425 pKa = 10.67HH426 pKa = 5.21LWNFLHH432 pKa = 6.58QKK434 pKa = 10.02EE435 pKa = 4.69EE436 pKa = 4.06YY437 pKa = 10.13LEE439 pKa = 4.19QVVGAYY445 pKa = 10.23LRR447 pKa = 11.84IYY449 pKa = 10.56ANSPQHH455 pKa = 4.98SQFWRR460 pKa = 11.84AVADD464 pKa = 3.79KK465 pKa = 11.25LNLRR469 pKa = 11.84VMSPSFYY476 pKa = 10.9SFFMNNPEE484 pKa = 3.94AAMKK488 pKa = 10.1LGPVRR493 pKa = 11.84YY494 pKa = 9.36RR495 pKa = 11.84EE496 pKa = 4.08GLLGGAGLVV505 pKa = 3.59
Molecular weight: 58.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
943 |
438 |
505 |
471.5 |
52.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.332 ± 0.765 | 1.379 ± 0.006 |
4.56 ± 0.444 | 5.302 ± 0.033 |
4.136 ± 0.764 | 6.681 ± 1.006 |
2.439 ± 0.047 | 3.287 ± 0.06 |
5.832 ± 0.679 | 9.756 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.439 ± 0.401 | 2.545 ± 0.426 |
6.469 ± 0.199 | 4.878 ± 0.502 |
5.726 ± 0.161 | 7.741 ± 1.059 |
4.772 ± 0.284 | 6.787 ± 1.086 |
2.121 ± 0.939 | 3.818 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
